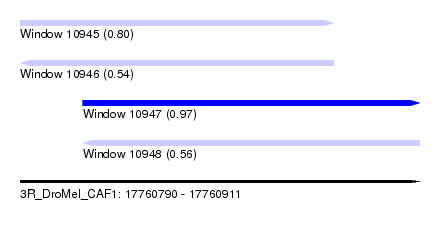
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,760,790 – 17,760,911 |
| Length | 121 |
| Max. P | 0.968871 |

| Location | 17,760,790 – 17,760,885 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -18.68 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
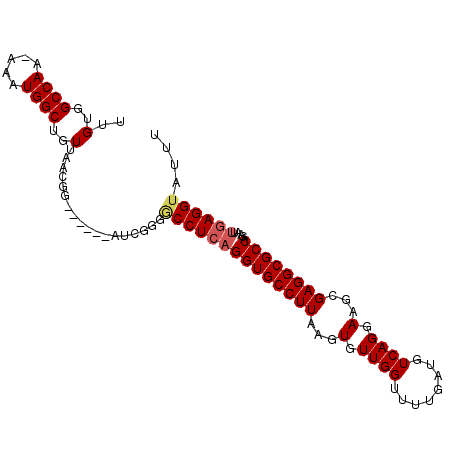
>3R_DroMel_CAF1 17760790 95 + 27905053 UAGGCGUCAGAAUGGACGGAAAUACCUCAAUUCUGGCGCCUCGCUUCCUGACAUCAAAACCAACACUUAAGGCACCUGAGGUCCCGAUUAACAGC .((((((((((((.((.((.....)))).)))))))))))).(((((..(((.(((...((.........))....))).)))..)).....))) ( -26.70) >DroSec_CAF1 49411 89 + 1 UAGGCGUCUGAAUGUACGGAAAUACCUCAAUUCUGGCGCCUCGCUUCCUGACAUGAAAACCAACACUUAAGGCACCUGAGGACCCGAU------C ..((((((.((((....((.....))...)))).))))))(((..((((.(..((....((.........))))..).))))..))).------. ( -21.30) >DroSim_CAF1 49829 89 + 1 UAGGCGUUUGAAUGUACGGAAAUACCUCAAUUCUGGCGCCUCGCUUCCUGACAUCAAAACCAACACUUAAGGCACCUGAGGCCCCGAU------C .((((((..((((....((.....))...))))..)))))).(((((.((....)).............((....)))))))......------. ( -18.90) >DroEre_CAF1 52126 89 + 1 UAAGCGCCAGAAUGCACGGAAAUACCUCCAUUCUGGCGCCUCGCUUCCUGACAUCAAAACCAACACUUAAGGCACCUGAGGCCCCGAU------C .....(((((((((...((.....))..)))))))))((((((...(((....................)))....))))))......------. ( -25.75) >DroYak_CAF1 53015 89 + 1 UAAGCGCCAGAAUGCACGGAAAUACCUGAAUUCUGGCGCCUCGCUUCCUGACAUCAAAACCAACACUUAAGGCACCUGAGGCCCCGAU------A .....((((((((.((.((.....)))).))))))))((((((...(((....................)))....))))))......------. ( -25.25) >consensus UAGGCGUCAGAAUGCACGGAAAUACCUCAAUUCUGGCGCCUCGCUUCCUGACAUCAAAACCAACACUUAAGGCACCUGAGGCCCCGAU______C .((((((((((((....((.....))...)))))))))))).(((((.((....)).............((....)))))))............. (-18.68 = -19.36 + 0.68)


| Location | 17,760,790 – 17,760,885 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -23.64 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17760790 95 - 27905053 GCUGUUAAUCGGGACCUCAGGUGCCUUAAGUGUUGGUUUUGAUGUCAGGAAGCGAGGCGCCAGAAUUGAGGUAUUUCCGUCCAUUCUGACGCCUA .(((...(((((((((...((..(.....)..)))))))))))..)))......(((((.((((((.((((.....)).)).)))))).))))). ( -31.70) >DroSec_CAF1 49411 89 - 1 G------AUCGGGUCCUCAGGUGCCUUAAGUGUUGGUUUUCAUGUCAGGAAGCGAGGCGCCAGAAUUGAGGUAUUUCCGUACAUUCAGACGCCUA (------(.((((.(((((((((((((........((((((......)))))))))))))).....)))))....)))).....))......... ( -25.80) >DroSim_CAF1 49829 89 - 1 G------AUCGGGGCCUCAGGUGCCUUAAGUGUUGGUUUUGAUGUCAGGAAGCGAGGCGCCAGAAUUGAGGUAUUUCCGUACAUUCAAACGCCUA .------....((((....((((((((...(.((((........)))).)...))))))))....(((((((((....)))).)))))..)))). ( -26.50) >DroEre_CAF1 52126 89 - 1 G------AUCGGGGCCUCAGGUGCCUUAAGUGUUGGUUUUGAUGUCAGGAAGCGAGGCGCCAGAAUGGAGGUAUUUCCGUGCAUUCUGGCGCUUA .------.((..(((.(((((.(((.........)))))))).)))..))....(((((((((((((.(((.....)).).))))))))))))). ( -34.60) >DroYak_CAF1 53015 89 - 1 U------AUCGGGGCCUCAGGUGCCUUAAGUGUUGGUUUUGAUGUCAGGAAGCGAGGCGCCAGAAUUCAGGUAUUUCCGUGCAUUCUGGCGCUUA .------.((..(((.(((((.(((.........)))))))).)))..))....((((((((((((.((((.....)).)).)))))))))))). ( -32.50) >consensus G______AUCGGGGCCUCAGGUGCCUUAAGUGUUGGUUUUGAUGUCAGGAAGCGAGGCGCCAGAAUUGAGGUAUUUCCGUACAUUCUGACGCCUA .........((((((((((((((((((...(.((((........)))).)...)))))))).....))))))...))))................ (-23.64 = -24.12 + 0.48)


| Location | 17,760,809 – 17,760,911 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -18.95 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.55 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17760809 102 + 27905053 AAAUACCUCAAUUCUGGCGCCUCGCUUCCUGACAUCAAAACCAACACUUAAGGCACCUGAGGUCCCGAUUAACAGCCGUUAACAGCCAUUUUUUGGCCACAA ....((((((.....((.((((.......((..........)).......)))).))))))))...(.(((((....)))))).((((.....))))..... ( -19.64) >DroSec_CAF1 49430 95 + 1 AAAUACCUCAAUUCUGGCGCCUCGCUUCCUGACAUGAAAACCAACACUUAAGGCACCUGAGGACCCGAU------CCGUUAACAGCCAUUU-UUGGCCACAA .....(((((.....((.((((.......((..........)).......)))).))))))).......------.........((((...-.))))..... ( -17.64) >DroSim_CAF1 49848 95 + 1 AAAUACCUCAAUUCUGGCGCCUCGCUUCCUGACAUCAAAACCAACACUUAAGGCACCUGAGGCCCCGAU------CCGUUAACAGCCAUUU-UUGGCCACAA ............((.((.((((((...(((....................)))....)))))))).)).------.........((((...-.))))..... ( -19.15) >DroEre_CAF1 52145 95 + 1 AAAUACCUCCAUUCUGGCGCCUCGCUUCCUGACAUCAAAACCAACACUUAAGGCACCUGAGGCCCCGAU------CCGUUAACAGCCAUUU-UUGGCCACAA ............((.((.((((((...(((....................)))....)))))))).)).------.........((((...-.))))..... ( -19.15) >DroYak_CAF1 53034 95 + 1 AAAUACCUGAAUUCUGGCGCCUCGCUUCCUGACAUCAAAACCAACACUUAAGGCACCUGAGGCCCCGAU------ACGUUAACAGCCAUUU-UUGGCCACAA ............((.((.((((((...(((....................)))....)))))))).)).------.........((((...-.))))..... ( -19.15) >consensus AAAUACCUCAAUUCUGGCGCCUCGCUUCCUGACAUCAAAACCAACACUUAAGGCACCUGAGGCCCCGAU______CCGUUAACAGCCAUUU_UUGGCCACAA ..............(((.((((((...(((....................)))....)))))).))).................((((.....))))..... (-17.47 = -17.55 + 0.08)


| Location | 17,760,809 – 17,760,911 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -26.72 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17760809 102 - 27905053 UUGUGGCCAAAAAAUGGCUGUUAACGGCUGUUAAUCGGGACCUCAGGUGCCUUAAGUGUUGGUUUUGAUGUCAGGAAGCGAGGCGCCAGAAUUGAGGUAUUU ......((....((((((((....)))))))).....))((((((((((((((...(.((((........)))).)...)))))))).....)))))).... ( -32.00) >DroSec_CAF1 49430 95 - 1 UUGUGGCCAA-AAAUGGCUGUUAACGG------AUCGGGUCCUCAGGUGCCUUAAGUGUUGGUUUUCAUGUCAGGAAGCGAGGCGCCAGAAUUGAGGUAUUU ..(..((((.-...))))..)......------...(((((((((((((((((........((((((......)))))))))))))).....))))).)))) ( -27.60) >DroSim_CAF1 49848 95 - 1 UUGUGGCCAA-AAAUGGCUGUUAACGG------AUCGGGGCCUCAGGUGCCUUAAGUGUUGGUUUUGAUGUCAGGAAGCGAGGCGCCAGAAUUGAGGUAUUU ..(..((((.-...))))..)......------......((((((((((((((...(.((((........)))).)...)))))))).....)))))).... ( -29.30) >DroEre_CAF1 52145 95 - 1 UUGUGGCCAA-AAAUGGCUGUUAACGG------AUCGGGGCCUCAGGUGCCUUAAGUGUUGGUUUUGAUGUCAGGAAGCGAGGCGCCAGAAUGGAGGUAUUU .((.((((..-...((((((....)))------.))).)))).))((((((((...(.((((........)))).)...))))))))............... ( -28.10) >DroYak_CAF1 53034 95 - 1 UUGUGGCCAA-AAAUGGCUGUUAACGU------AUCGGGGCCUCAGGUGCCUUAAGUGUUGGUUUUGAUGUCAGGAAGCGAGGCGCCAGAAUUCAGGUAUUU ..(..((((.-...))))..)....((------(((..((..((.((((((((...(.((((........)))).)...)))))))).))..)).))))).. ( -26.40) >consensus UUGUGGCCAA_AAAUGGCUGUUAACGG______AUCGGGGCCUCAGGUGCCUUAAGUGUUGGUUUUGAUGUCAGGAAGCGAGGCGCCAGAAUUGAGGUAUUU ..(..((((.....))))..)..................((((((((((((((...(.((((........)))).)...)))))))).....)))))).... (-26.72 = -27.20 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:48 2006