
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,641,914 – 17,642,008 |
| Length | 94 |
| Max. P | 0.800275 |

| Location | 17,641,914 – 17,642,008 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.13 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
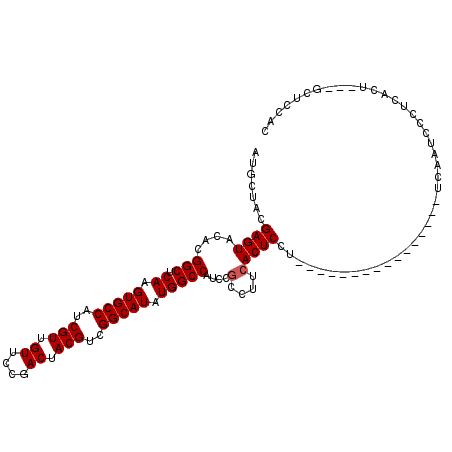
>3R_DroMel_CAF1 17641914 94 - 27905053 AUGCUACGAGUACAUGGCUCAAGUGCCAUCGUUGUUCCGACUACGUCGGCAUAUGGCCAUCCGCCUUCACUCCU----------------UCAAUCCCUCACU---GCUCCAC .......(((((.(((((.((.(((((..(((.((....)).)))..))))).)))))))..(......)....----------------............)---))))... ( -22.30) >DroSec_CAF1 36729 94 - 1 AUGCUACGAGUACACGGCUCAAGUGCCAUCGUUGUUCCGACUACGUCGGCAUAUGGCCAUCCGCCUUCACUCCU----------------UCAAUCCCUCACU---GAUCCAC .......((((....(((.((.(((((..(((.((....)).)))..))))).)))))....(....)))))..----------------.............---....... ( -20.00) >DroSim_CAF1 36555 94 - 1 AUGCUACGAGUACACGGCUCAAGUGCCAUCGUUGUUCCGACUACGUCGGCAUAUGGCCAUCCGCCUUCACUCCU----------------UCAAUCCCUCACU---GCUCCAC .......(((((...(((.((.(((((..(((.((....)).)))..))))).)))))....(......)....----------------............)---))))... ( -21.20) >DroEre_CAF1 38491 102 - 1 AUGCUACGAGUACACGGCUCAAGUGCCAUCGUUGUUCCGACUACGUCGGCAUAUGGCCAUCCGUCUUCACUCCUUCAC--------UCCUCCAAUGCCCCACU---GCUCCCC .......(((((...(((.((.(((((..(((.((....)).)))..))))).)))))....((....))........--------................)---))))... ( -22.20) >DroYak_CAF1 41875 110 - 1 AUGCUACGAGUACACGGCUCAAGUGCCAUCGUUGUUCCGACUACGUCGGCAUAUGGCCAUCCGCCUUCACUCCUUUACUCCAUCGCUCCUUCAAUCCCUCACU---GCUCCAC .......(((((...(((.((.(((((..(((.((....)).)))..))))).)))))....((....................))................)---))))... ( -22.55) >DroAna_CAF1 45052 94 - 1 AUGCUACGAGUACACGGCUCAAGUGCCAUCGUUGUUCCGACUACGUCGGCAUAUGGCCAUCCUUUUUCACUCU---AC----------------UCCUUCCCUCCACCUCCAC .......(((((...(((.((.(((((..(((.((....)).)))..))))).)))))..............)---))----------------))................. ( -19.73) >consensus AUGCUACGAGUACACGGCUCAAGUGCCAUCGUUGUUCCGACUACGUCGGCAUAUGGCCAUCCGCCUUCACUCCU________________UCAAUCCCUCACU___GCUCCAC .......((((....(((.((.(((((..(((.((....)).)))..))))).)))))....(....)))))......................................... (-19.52 = -19.68 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:04 2006