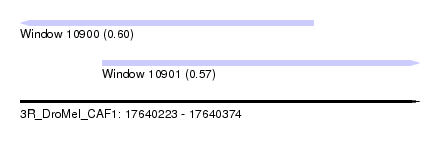
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,640,223 – 17,640,374 |
| Length | 151 |
| Max. P | 0.602843 |

| Location | 17,640,223 – 17,640,334 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -18.26 |
| Energy contribution | -18.27 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17640223 111 - 27905053 UUGGGAUCAAAGUGCUUUCAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUUGUAAUAUAUAUACAAUAUUUUAAAUGAAAAAGUAUAGGAAAUCAAAAACAAAAGUG (((((.(((((((((((....))))...)))))))))(((((((.((...((((((.(((((.......))))).))))))...)).))))))).........)))..... ( -18.80) >DroSec_CAF1 35044 109 - 1 UCGGGAUCAAAGUGGUUCGAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUUGUAAUAU--AUACACUAUUUUAAAUGAAGAAGUAUGUGCAAUCAAAACAAAAAUUU ..(((.(((((((((.((....))..))))))))))))......(((((((((..(((....--.)))(((.((((....)))).)))))))))))).............. ( -25.40) >DroSim_CAF1 34866 109 - 1 UCGGGAUCAAAGUGCUUCGAAAGGCUUCACUUUGACCUUUUAAAGUUGCAUAUUUGUAAUAU--AUACAAUAUUUUAAAUGAAGAAGUAUAUGCAAUCAAAACAAAAAUUU ..(((.((((((((..((....))...)))))))))))......((((((((.(((((....--.)))))((((((.......)))))))))))))).............. ( -24.50) >consensus UCGGGAUCAAAGUGCUUCGAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUUGUAAUAU__AUACAAUAUUUUAAAUGAAGAAGUAUAUGCAAUCAAAACAAAAAUUU ..(((.((((((((..((....))...)))))))))))......((((((((..((((.......)))).((((((.......)))))))))))))).............. (-18.26 = -18.27 + 0.01)



| Location | 17,640,254 – 17,640,374 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.53 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -20.25 |
| Energy contribution | -20.25 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17640254 120 + 27905053 AUUUAAAAUAUUGUAUAUAUAUUACAAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUGAAAGCACUUUGAUCCCAACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUU ............((((((.(.((((..................((((((((((...(....)....)))))))))).....(((((.((....)).)))))...)))).).))))))... ( -22.50) >DroSec_CAF1 35075 118 + 1 AUUUAAAAUAGUGUAU--AUAUUACAAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUCGAACCACUUUGAUCCCGACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUU .......(((((((((--((.......)))))).)))))....((((((((((.............)))))))))).....(.(((((.(((((..........)))))..))))).).. ( -23.62) >DroSim_CAF1 34897 118 + 1 AUUUAAAAUAUUGUAU--AUAUUACAAAUAUGCAACUUUAAAAGGUCAAAGUGAAGCCUUUCGAAGCACUUUGAUCCCGACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUU ...........(((((--((.......)))))))(((((....((((((((((...(.....)...)))))))))).....(((((.((....)).)))))))))).............. ( -23.90) >consensus AUUUAAAAUAUUGUAU__AUAUUACAAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUCGAAGCACUUUGAUCCCGACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUU ............((((.....((((..................((((((((((.............)))))))))).....(((((.((....)).)))))...)))).....))))... (-20.25 = -20.25 + -0.00)

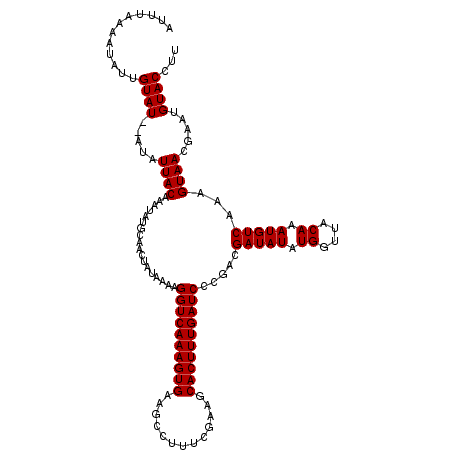

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:03 2006