
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,638,512 – 17,638,639 |
| Length | 127 |
| Max. P | 0.996511 |

| Location | 17,638,512 – 17,638,602 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -10.34 |
| Energy contribution | -11.15 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17638512 90 - 27905053 CAC----------CACCCGCAUGAUGCGUUGGAAAACUGACGAUUAUGGGGGAGCUGGGCUG--CUAGACCACUUAAUUACUUUUCAAUUUAAAUCAAAUUA ...----------..(((.((((((.(((..(....)..))))))))).)))(((......)--)).................................... ( -21.40) >DroSec_CAF1 33390 90 - 1 CUC----------CACCCGCAUGAUGCGUUGGAAAACUGACGAUUAUGUGGGAGAUGGGCUG--CUAGACCACUUAAUUACUUUUCAAUUUAAAUCAAAUUA ...----------..((((((((((.(((..(....)..)))))))))))))((.(((....--.....)))))............................ ( -22.80) >DroSim_CAF1 33207 90 - 1 CUC----------CACCCGCAUGAUGCGUUGGAAAACUGACGAUUAUGUGGGAGAUGGGCUG--CUAGACCACUUAAUUACUUUUCAAUUUAAAUCAAAUUA ...----------..((((((((((.(((..(....)..)))))))))))))((.(((....--.....)))))............................ ( -22.80) >DroEre_CAF1 35251 82 - 1 CAG----------CACCCGCAUAAUGCGUUCCAAAAGUGACGAUUAUGGGGGAGAUGG----------ACCACUUAAUUACUUUUCAAUUUAAGUCAAAUUA ((.----------(.(((.((((((.(((..(....)..))))))))).))).).)).----------...((((((............))))))....... ( -18.40) >DroYak_CAF1 38148 93 - 1 CAC-------CAUCACCCGCAUAAUGCGUUCAAAAACUGACGAUUAUGGGGGAGAUGGACUG--ACAGACCACUUAAUUACUUUUCAAUUUAAAUCAAAUUA ..(-------((((.(((.((((((.((((........)))))))))).))).)))))....--...................................... ( -23.80) >DroAna_CAF1 41862 94 - 1 CAGGAAAAUACGCUUCCCGCAUAAAGCUUUACAG---GGAGAACUACUCCACA-----GCUACUCCACAACACUUAAUUAGUUUUCCGUUUAAAUCAAAUAU ..(((((((..((.....))....((((......---((((.....))))..)-----)))...................)))))))............... ( -16.30) >consensus CAC__________CACCCGCAUAAUGCGUUGGAAAACUGACGAUUAUGGGGGAGAUGGGCUG__CUAGACCACUUAAUUACUUUUCAAUUUAAAUCAAAUUA ...............(((.((((((.((((........)))))))))).))).................................................. (-10.34 = -11.15 + 0.81)

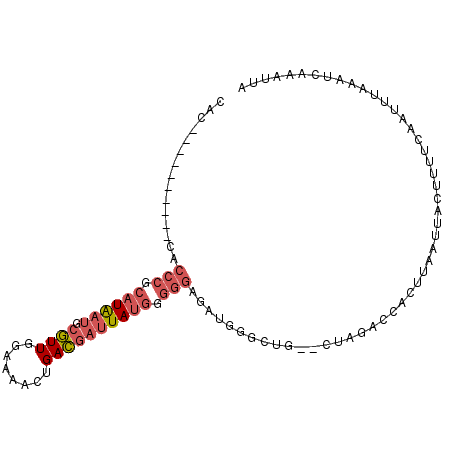
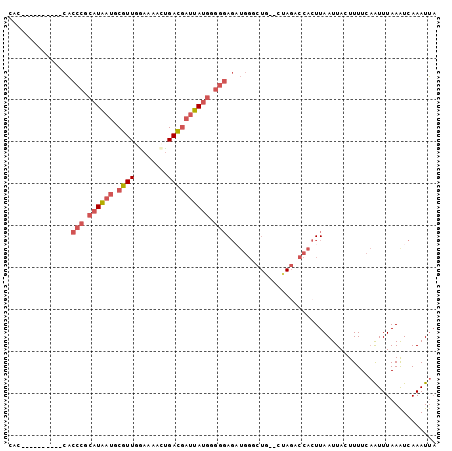
| Location | 17,638,534 – 17,638,639 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.91 |
| Mean single sequence MFE | -32.96 |
| Consensus MFE | -20.29 |
| Energy contribution | -21.16 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17638534 105 - 27905053 AAUGCCCGCUGAA-AUGGGUGGG-GAAAAACUAGGAGAACAC---CACCCGCAUGAUGCGUUGGAAAACUGACGAUUAUGGGGGAGCUGGGCUGCUAGACCACUUAAUUA ..(.((((((...-...))))))-.)....((((.((..((.---(.(((.((((((.(((..(....)..))))))))).))).).))..)).))))............ ( -38.90) >DroSec_CAF1 33412 106 - 1 AAUGCCCGCUGAA-GUGGGUGGGGGGAAAACUAGGAGAGCUC---CACCCGCAUGAUGCGUUGGAAAACUGACGAUUAUGUGGGAGAUGGGCUGCUAGACCACUUAAUUA ...((((((....-))))))(((((.....((((...(((.(---((((((((((((.(((..(....)..)))))))))))))...)))))).)))).)).)))..... ( -40.50) >DroSim_CAF1 33229 106 - 1 AAUGCCCGCUGAA-GUGGGUGGGGGAAAAACUAGGAGAGCUC---CACCCGCAUGAUGCGUUGGAAAACUGACGAUUAUGUGGGAGAUGGGCUGCUAGACCACUUAAUUA ...((((((....-))))))(((((.....((((...(((.(---((((((((((((.(((..(....)..)))))))))))))...)))))).)))).)).)))..... ( -39.40) >DroEre_CAF1 35273 79 - 1 AA-------------UG-------UGAAAACUAGGAGCGCAG---CACCCGCAUAAUGCGUUCCAAAAGUGACGAUUAUGGGGGAGAUGG--------ACCACUUAAUUA ..-------------.(-------(....))..((..(.((.---(.(((.((((((.(((..(....)..))))))))).))).).)))--------.))......... ( -19.20) >DroYak_CAF1 38170 98 - 1 AAUG-----UGAAAAUG-------UGAAAACUAGGAGAGCACCAUCACCCGCAUAAUGCGUUCAAAAACUGACGAUUAUGGGGGAGAUGGACUGACAGACCACUUAAUUA ...(-----((.....(-------(....))..........(((((.(((.((((((.((((........)))))))))).))).)))))..........)))....... ( -26.80) >consensus AAUGCCCGCUGAA_AUGGGUGGG_GGAAAACUAGGAGAGCAC___CACCCGCAUGAUGCGUUGGAAAACUGACGAUUAUGGGGGAGAUGGGCUGCUAGACCACUUAAUUA ....((((((.......))))))........((((............(((.((((((.((((((....)))))))))))).)))...(((.........))))))).... (-20.29 = -21.16 + 0.87)

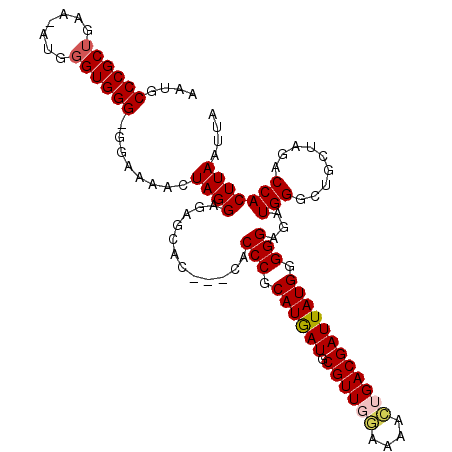

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:01 2006