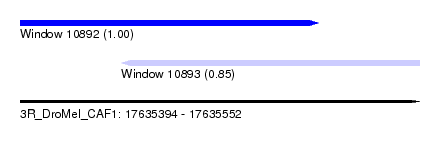
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,635,394 – 17,635,552 |
| Length | 158 |
| Max. P | 0.995226 |

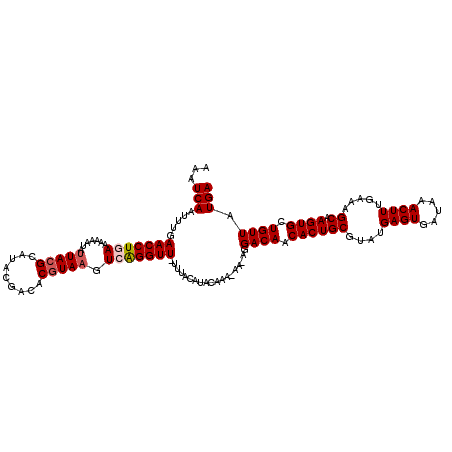
| Location | 17,635,394 – 17,635,512 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.70 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.47 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17635394 118 + 27905053 AUAAAAACAGCACUUUUGGUGUAAGUAAGUCAUAAAAAACUCAUAACAGCACUUGCUUUCAAAGUUUAUCACUCAUACGCAGUGUUGUCCU-UUUUUUGUAUGUAAA-AACCUGACUUAU .........((((.....))))..((((((((.............((((((((.((......(((.....))).....))))))))))...-.((((((....))))-))..)))))))) ( -21.30) >DroSec_CAF1 30283 117 + 1 AUAAAAACAGCACUUUUGGUGUAAGUAAGUCAUAAAAAACUCAUAACAGCACUUGCUUUCAAAGUUUAUCACUCAUACGCAGUGUUGUCCU-UU-UUUGUAUGUAAA-AACCUGACUUAC .........((((.....))))..((((((((.........((((((((((((.((......(((.....))).....))))))))))...-..-....))))....-....)))))))) ( -22.53) >DroSim_CAF1 30092 117 + 1 AUAAAAACAGCACUUUUGGUGUAAGUAAGUCAUAAAAAACUCAUAACAGCACUUGCUUUCAAAGUUUAUCACUCAUACGCAGUGUUGUCCU-UU-UUCGUAUGUAAA-AACCUGACUUAC .........((((.....))))..((((((((.........((((((((((((.((......(((.....))).....))))))))))...-..-....))))....-....)))))))) ( -22.53) >DroEre_CAF1 32190 117 + 1 AUAAAAACAGCACUUUUGGUGUAAGUAAGUCAUAAAAAACUCAUAACAGCACUUGCUUUCAAAGUUUAUCACUCAUACGCAGUGUUGUCCU-UU-UUGGUAUGUAAA-AACCUGACUUAC .........((((.....))))..((((((((.........((((((((((((.((......(((.....))).....))))))))))((.-..-..))))))....-....)))))))) ( -24.53) >DroYak_CAF1 34801 119 + 1 AUAAAAACAGCACUUUCGGUGUAAGUAAGUCAUAAAAAACUCAUAACAACACUUGCUUUCAAAGUUUAUCACUCAUACGCAGUGUUGUCCUGUU-UUUGUAUGUAAAAAACCCGACAUAC ..(((((((((((.....)))........................((((((((.((......(((.....))).....)))))))))).)))))-)))((((((..........)))))) ( -21.20) >consensus AUAAAAACAGCACUUUUGGUGUAAGUAAGUCAUAAAAAACUCAUAACAGCACUUGCUUUCAAAGUUUAUCACUCAUACGCAGUGUUGUCCU_UU_UUUGUAUGUAAA_AACCUGACUUAC .........((((.....))))..((((((((.........((((((((((((.((......(((.....))).....))))))))))...........)))).........)))))))) (-21.39 = -21.47 + 0.08)

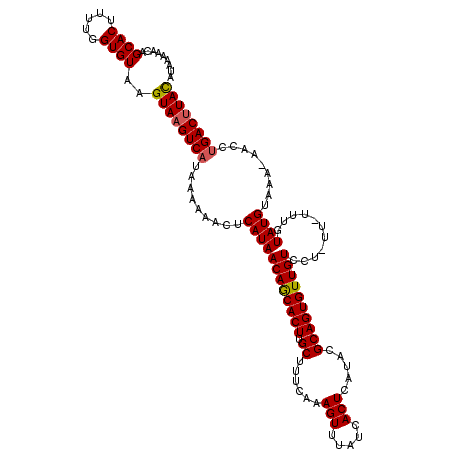
| Location | 17,635,434 – 17,635,552 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -19.44 |
| Energy contribution | -20.08 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17635434 118 - 27905053 AAAUCAAUUUGAACCUGAAAAAAUAUUACGAAUACGACACAUAAGUCAGGUU-UUUACAUACAAAAAA-AGGACAACACUGCGUAUGAGUGAUAAACUUUGAAAGCAAGUGCUGUUAUGA ...(((....((((((((..........((....)).........)))))))-)..............-..((((.((((((....((((.....)))).....)).)))).)))).))) ( -21.81) >DroSec_CAF1 30323 117 - 1 AAAUCAAUUUGAACCUGAAAAAAUAUUACGCAUACGACACGUAAGUCAGGUU-UUUACAUACAAA-AA-AGGACAACACUGCGUAUGAGUGAUAAACUUUGAAAGCAAGUGCUGUUAUGA ...(((....((((((((.......(((((.........))))).)))))))-)...........-..-..((((.((((((....((((.....)))).....)).)))).)))).))) ( -24.20) >DroSim_CAF1 30132 117 - 1 AAAUCAAUUUGAACCUGAAAAAAUAUUACGCAUACGACACGUAAGUCAGGUU-UUUACAUACGAA-AA-AGGACAACACUGCGUAUGAGUGAUAAACUUUGAAAGCAAGUGCUGUUAUGA ...(((....((((((((.......(((((.........))))).)))))))-)...........-..-..((((.((((((....((((.....)))).....)).)))).)))).))) ( -24.20) >DroEre_CAF1 32230 117 - 1 AAAUCAAUUUGAACCUAAAAGAAUAUUACGCAUACGACACGUAAGUCAGGUU-UUUACAUACCAA-AA-AGGACAACACUGCGUAUGAGUGAUAAACUUUGAAAGCAAGUGCUGUUAUGA ......(((((......((((..((((((.((((((.((.((..(((.(((.-.......)))..-..-..)))...)))))))))).))))))..)))).....))))).......... ( -22.70) >DroYak_CAF1 34841 119 - 1 AAAUCAAUUUGAACCUAAAAGAAUAUUACGCAUACGACACGUAUGUCGGGUUUUUUACAUACAAA-AACAGGACAACACUGCGUAUGAGUGAUAAACUUUGAAAGCAAGUGUUGUUAUGA .......((((......(((((((....((((((((...)))))).)).))))))).....))))-..((.(((((((((((....((((.....)))).....)).))))))))).)). ( -28.70) >consensus AAAUCAAUUUGAACCUGAAAAAAUAUUACGCAUACGACACGUAAGUCAGGUU_UUUACAUACAAA_AA_AGGACAACACUGCGUAUGAGUGAUAAACUUUGAAAGCAAGUGCUGUUAUGA ...(((.....(((((((.......(((((.........))))).)))))))...................((((.((((((....((((.....)))).....)).)))).)))).))) (-19.44 = -20.08 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:56 2006