
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,631,617 – 17,631,742 |
| Length | 125 |
| Max. P | 0.962486 |

| Location | 17,631,617 – 17,631,710 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.57 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17631617 93 + 27905053 -----------AGCGAAACAGAAC-UUGGUGGGCUGGCAGGCGAAGGCGGAACGGG--GCGUUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGCCU -----------..((...(((..(-.....)..)))....((....))....))((--((.((((..((((((((((((....)).))))))))))..)))).)))) ( -30.10) >DroSec_CAF1 26537 93 + 1 -----------AGCGAAACAGAAC-UUGGUGGGCUGGCAGGCGAAGGCGGAGCAGG--GCUUUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGUCU -----------.((....(((..(-.....)..)))....((....))...)).((--(((((((..((((((((((((....)).))))))))))..))))))))) ( -33.10) >DroSim_CAF1 26363 93 + 1 -----------AGCGAAACAGAAC-UUGGUGGGCUGGCAGGCGAAGGCGGAGUAGG--GCUUUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGCCU -----------.((....(((..(-.....)..)))....((....))...)).((--(((((((..((((((((((((....)).))))))))))..))))))))) ( -33.50) >DroEre_CAF1 28353 94 + 1 ----------AAGCGAAACAGAAG-UUGGUGGGCUGGCAGGCGGAGGCGGAACAGG--GCUUUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGCCU ----------..............-....((..(((.(.......).)))..))((--(((((((..((((((((((((....)).))))))))))..))))))))) ( -31.00) >DroYak_CAF1 30884 91 + 1 CCGAAACCGAAACCGAAACAGAA--------------CAGGCGAAGGCGGAACGGG--GCUGUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGCCU ......(((...((....(....--------------..)((....))))..)))(--(((.(((..((((((((((((....)).))))))))))..))).)))). ( -28.50) >DroAna_CAF1 34918 94 + 1 -----------A--AACACAAAACCUUCGGGGAGUCGGCGGCGCGGGCGGAACGGAGCGCUUUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGCCU -----------.--..........(((((....(((.((...)).)))....))))).(((((((..((((((((((((....)).))))))))))..))))))).. ( -32.20) >consensus ___________AGCGAAACAGAAC_UUGGUGGGCUGGCAGGCGAAGGCGGAACAGG__GCUUUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGCCU ........................................((....))..........(((((((..((((((((((((....)).))))))))))..))))))).. (-22.37 = -22.57 + 0.20)


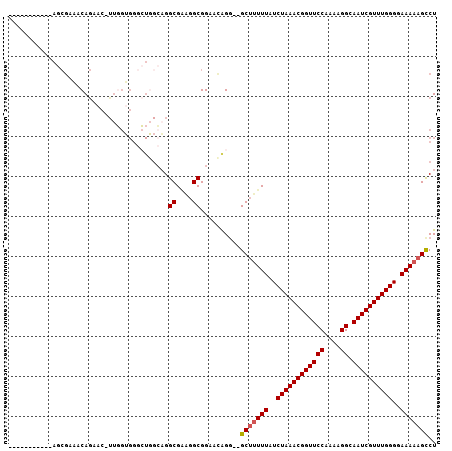
| Location | 17,631,645 – 17,631,742 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.92 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
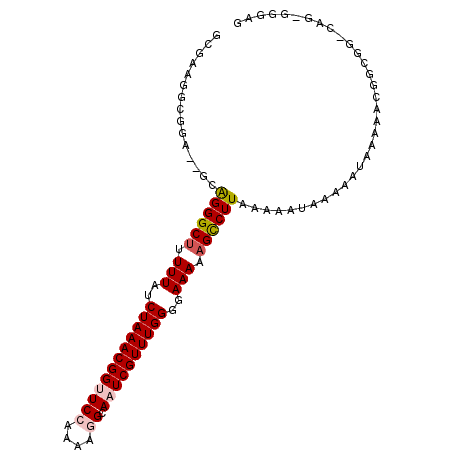
>3R_DroMel_CAF1 17631645 97 + 27905053 GCGAAGGCGGA--ACGGGGCGUUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGCCUUAAAAAUAAAAAUAAAAACGGCGG-CAG-GGGAG ((....))...--..(((((.((((..((((((((((((....)).))))))))))..)))).))))).......................-...-..... ( -26.00) >DroPse_CAF1 22801 82 + 1 AG--AUACGGAGAGGAGGGCU-UUUAUCUAAACGGCUCCAAACAGCAAUCGUUUGGGGAAAAAGCCUUAAAAAUGUAAAUAAAAA---------------- ..--.((((......((((((-(((.(((((((((((......)))...))))))))..))))))))).....))))........---------------- ( -22.90) >DroSec_CAF1 26565 99 + 1 GCGAAGGCGGA--GCAGGGCUUUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGUCUUAAAAAUAAAAAUAAAAACGGCGGGCAGGGGGAG ((....))...--((((((((((((..((((((((((((....)).))))))))))..)))))))))).................(....)))........ ( -27.70) >DroSim_CAF1 26391 97 + 1 GCGAAGGCGGA--GUAGGGCUUUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGCCUUAAAAAUAAAAAUAAAAACGGCGG-CAG-GGGAG ((....))...--.(((((((((((..((((((((((((....)).))))))))))..)))))))))))......................-...-..... ( -31.50) >DroYak_CAF1 30910 97 + 1 GCGAAGGCGGA--ACGGGGCUGUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGCCUUAAAAAUAAAAAUAAAAACGGCGG-CAG-GGGGA ((....))...--..((((((.(((..((((((((((((....)).))))))))))..))).)))))).......................-...-..... ( -26.60) >DroPer_CAF1 17895 80 + 1 AG--AUACGGA--GGAGGGCU-UUUAUCUAAACGGCUCCAAACAGCAAUCGUUUGGGGAAAAAGCCUUAAAAAUGUAAAUAAAAA---------------- ..--.((((..--..((((((-(((.(((((((((((......)))...))))))))..))))))))).....))))........---------------- ( -23.10) >consensus GCGAAGGCGGA__GCAGGGCUUUUUAUCUAAACGGUUCCAAAAGGCAAUCGUUUGGGGAAAAAGCCUUAAAAAUAAAAAUAAAAACGGCGG_CAG_GGGAG ...............((((((.(((..((((((((((((....)).))))))))))..))).))))))................................. (-19.44 = -19.92 + 0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:53 2006