
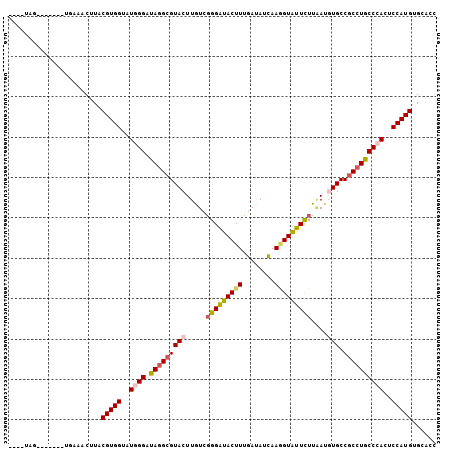
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,613,487 – 17,613,589 |
| Length | 102 |
| Max. P | 0.961608 |

| Location | 17,613,487 – 17,613,589 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -27.57 |
| Energy contribution | -27.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17613487 102 + 27905053 --ACUAGAAUGU--AGAAACUUACGUGGUAUGGGAUAGGCGUACUUGUCGGGGUACUUCGAUAUCAAGGUGUUCUUAAUGUGCCGCCUGCCCACCCCAUGAGCACC --..........--.........(((((..((((.(((((((((...(.((..(((((.(....).)))))..)).)..))).))))))))))..)))))...... ( -34.30) >DroVir_CAF1 12650 95 + 1 ----UAG-------UUUAACUUACGUGGUAUGGGAUAGGCGUACUUAUCGGGAUACUUUGAUAUUAAAGUAUUCUUAAUAUGCCGCCUGCCCACGCCAUGUGCGCC ----...-------.......(((((((..((((.((((((........(((((((((((....)))))))))))........))))))))))..))))))).... ( -37.89) >DroGri_CAF1 10096 95 + 1 ----UCG-------UGUCACUUACGUGGUAUGGGAUAGGCGUACUUAUCGGGAUACUUUGAUAUUAAAGUAUUCUUUAUGUGCCUCCUACCAACUCCAUGUGCACC ----..(-------(((.....((((((....((.((((.((((.....(((((((((((....)))))))))))....))))..))))))....)))))))))). ( -30.90) >DroYak_CAF1 8711 94 + 1 ----------AU--AUCAACUUACGUGGUAUGGGAUAGGCGUACUUGUCGGGAUACUUCGAUAUCAAGGUGUUCUUGAUGUGCCGCCUGCCGACUCCAUGAGCACC ----------..--.........(((((..(.((.(((((((((..((((((((((((.(....).))))).)))))))))).)))))))).)..)))))...... ( -31.80) >DroMoj_CAF1 9805 94 + 1 ----UAG-------UU-GACUUACGUGGUAUGGGAUAAGCGUACUUGUCGGGAUACUUUGAUAUCAAUGUAUUUUUAAUAUGCCGCCUACCCACGCCAUGUGCGCC ----...-------..-....(((((((..((((.((.(((......(..((((((.(((....))).))))))..)......))).))))))..))))))).... ( -27.80) >DroAna_CAF1 17387 106 + 1 UCGGCAGAAUGCCAUGAAACUUACGUGGAAUGGGAUAGGCGUACUUGUCCGGGUACUUGGAAAUCAAGGUGUUCUUGAUGUGCCGCCUGCCCACUCCAUGGGCACC ..(((.....)))..........((((((.((((.(((((((((((....))))))..((..((((((.....))))))...))))))))))).))))))...... ( -44.00) >consensus ____UAG_______UGAAACUUACGUGGUAUGGGAUAGGCGUACUUGUCGGGAUACUUUGAUAUCAAGGUAUUCUUAAUGUGCCGCCUGCCCACUCCAUGUGCACC .......................(((((..((((.(((((((((.....(((((((((........)))))))))....))).))))))))))..)))))...... (-27.57 = -27.90 + 0.33)


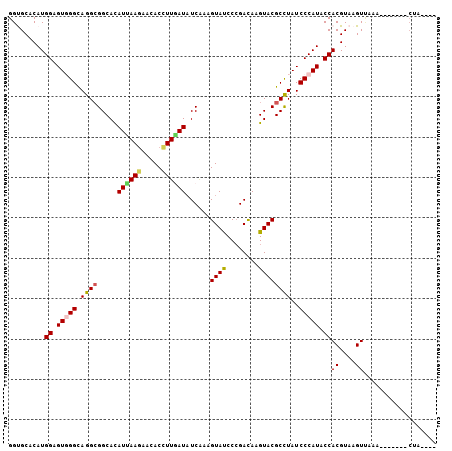
| Location | 17,613,487 – 17,613,589 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -17.64 |
| Energy contribution | -17.67 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17613487 102 - 27905053 GGUGCUCAUGGGGUGGGCAGGCGGCACAUUAAGAACACCUUGAUAUCGAAGUACCCCGACAAGUACGCCUAUCCCAUACCACGUAAGUUUCU--ACAUUCUAGU-- .(((...(((((((((((...(((...((((((.....)))))).)))..((((........)))))))))))))))..)))(((......)--))........-- ( -29.90) >DroVir_CAF1 12650 95 - 1 GGCGCACAUGGCGUGGGCAGGCGGCAUAUUAAGAAUACUUUAAUAUCAAAGUAUCCCGAUAAGUACGCCUAUCCCAUACCACGUAAGUUAAA-------CUA---- (((..((.(((..((((.(((((((.((((..((.((((((......))))))))..)))).)).)))))..))))..))).))..)))...-------...---- ( -29.00) >DroGri_CAF1 10096 95 - 1 GGUGCACAUGGAGUUGGUAGGAGGCACAUAAAGAAUACUUUAAUAUCAAAGUAUCCCGAUAAGUACGCCUAUCCCAUACCACGUAAGUGACA-------CGA---- .((((((.((..((.((((((((((...(((((....)))))........((((........))))))))...)).)))))).)).))).))-------)..---- ( -22.30) >DroYak_CAF1 8711 94 - 1 GGUGCUCAUGGAGUCGGCAGGCGGCACAUCAAGAACACCUUGAUAUCGAAGUAUCCCGACAAGUACGCCUAUCCCAUACCACGUAAGUUGAU--AU---------- (((....((((....(..(((((..((((((((.....)))))).(((........)))...)).)))))..))))))))((....))....--..---------- ( -22.10) >DroMoj_CAF1 9805 94 - 1 GGCGCACAUGGCGUGGGUAGGCGGCAUAUUAAAAAUACAUUGAUAUCAAAGUAUCCCGACAAGUACGCUUAUCCCAUACCACGUAAGUC-AA-------CUA---- (((..((.(((..((((((((((...........((((.(((....))).))))...........)))))).))))..))).))..)))-..-------...---- ( -28.65) >DroAna_CAF1 17387 106 - 1 GGUGCCCAUGGAGUGGGCAGGCGGCACAUCAAGAACACCUUGAUUUCCAAGUACCCGGACAAGUACGCCUAUCCCAUUCCACGUAAGUUUCAUGGCAUUCUGCCGA ((((((...((((((((.((((((...((((((.....))))))..))..((((........))))))))..))))))))((....)).....))))))....... ( -38.30) >consensus GGUGCACAUGGAGUGGGCAGGCGGCACAUUAAGAACACCUUGAUAUCAAAGUAUCCCGACAAGUACGCCUAUCCCAUACCACGUAAGUUAAA_______CUA____ .........((.(((((.((((.....((((((.....))))))......((((........))))))))..))))).))((....)).................. (-17.64 = -17.67 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:45 2006