
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,603,353 – 17,603,460 |
| Length | 107 |
| Max. P | 0.648176 |

| Location | 17,603,353 – 17,603,460 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -19.52 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
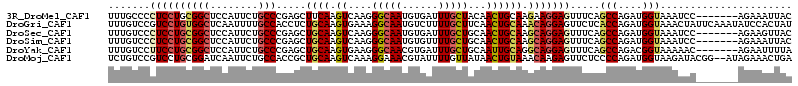
>3R_DroMel_CAF1 17603353 107 - 27905053 UUUGCCCCUCCUGCGGCUCCAUUCUGCCCGAGCUUCAAGUCAAGGGCAAUGUGAUUUGCUACAACUGCAAGAAGGAGUUUCAGCCAGAUGGUAAAUCC-------AGAAAUUAC ((((((.((.(((..(((((.((((((((..((.....))...))))).......((((.......))))))))))))..)))..))..))))))...-------......... ( -30.30) >DroGri_CAF1 7589 114 - 1 UUUGUCCGUCCUGUGGCUCAAUUUUGCCACCUCUGCAAGUGAAAGGCAAUGUCUUUUGCUUCAACUGCAAACAGGAGUUCUCACCAGAUGGUAAACUAUUCAAAUAUCCACUAU ((((.(((((..(((((........)))))(((((.(((..((((((...))))))..))))).(((....)))))).........)))))))))................... ( -28.30) >DroSec_CAF1 5796 107 - 1 UUUGUCCCUCCUGCGGCUCCAUUCUGCCCGAGCUGCAAGUCAAGGGCAAUGUGAUUUGCUGCAACUGCAAGCAGGAGUUUCAGCCAGAUGGUAAAUCC-------AGAAGUUAC .((((((..(.((((((((..........)))))))).)....)))))).(((((((((((.((((.(.....).)))).))))....(((.....))-------).))))))) ( -33.20) >DroSim_CAF1 5734 107 - 1 UUUGUCCCUCCUGCGGCUCCAUUCUGCCCGAGCUGCAAGUCAAGGGCAAUGUGUUUUGCUGCAACUGCAAGCAGGAGUUUCAGCCAGAUGGUAAAUCC-------AGAAAUUAC ((((...((((((((((..(((..(((((..((.....))...)))))..)))....)))((....))..))))))).....(((....))).....)-------)))...... ( -32.20) >DroYak_CAF1 5716 107 - 1 UUUGUCCUUCCUGCGGCUCCAUUCUGCCCGAGCUGCAAGUGAAGGGCAACGUGAUUUGCUGCAAUUGCAGGCAGGAGUUUCAGCCAGACGGUAAAAAC-------AGAAUUUUA .((((((((((((((((((..........))))))).)).))))))))).....((((((((....))).))))).((((..(((....)))..))))-------......... ( -37.50) >DroMoj_CAF1 6026 112 - 1 UCUGUCCGUCCUGCGGAUCAAUUCUGCCACCGCUGCAAGUCAAAGGAAACGUAUUUUGUUAUAACUGUAAACAAGAGUUCUCCCCAGAUGGUAAGAUACGG--AUAGAAACUGA (((((((((...(((((.....))))).(((((((.........(....)...(((((((.........)))))))........))).)))).....))))--)))))...... ( -29.10) >consensus UUUGUCCCUCCUGCGGCUCCAUUCUGCCCGAGCUGCAAGUCAAGGGCAAUGUGAUUUGCUGCAACUGCAAGCAGGAGUUUCAGCCAGAUGGUAAAUCC_______AGAAAUUAC .......((((((((((........))).....((((.((....(((((......)))))...)))))).))))))).....(((....)))...................... (-19.52 = -20.22 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:42 2006