
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,577,128 – 17,577,231 |
| Length | 103 |
| Max. P | 0.925309 |

| Location | 17,577,128 – 17,577,231 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.55 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -18.13 |
| Energy contribution | -19.13 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17577128 103 + 27905053 UA---AUUUG-----UAAUU-------UCGAUUACAGGCCUAUCCCGAGUGGGACAUUUUCCAGACCUUCCACGCUGGAUCGUACCAGAGCGCUCUGGUGAAUGAUGCCAGCGCUAAU ..---...((-----((((.-------...))))))(((.........((((((..((.....))..))))))((((((((((((((((....))))))..))))).))))))))... ( -33.60) >DroVir_CAF1 15391 107 + 1 UA---ACUUC--AAUUGAUUU------UCUGUCCAAGGCCUAUCCAGAAUGGGAUAUAUUCCAAACCUUCCAUGCUGGCUCUUAUCAGAGCGCUCUCAUUGCUGAUGCCAACCCCAAU ..---.....--.........------.........(((..((((((.((((((.............)))))).)))(((((....)))))((.......)).))))))......... ( -19.32) >DroPse_CAF1 12309 110 + 1 AG---CUUUA-----UAAUUAUACCAAUCUAUUGCAGGCCUAUCCGGAAUGGGACAUUUUCCAGAUCUUCCAUGCCGGCUCCUACCAAAGCGCCCUGAUCAACGACGCCACUCCCACU .(---((((.-----.........(((....))).(((.....((((.((((((.(((.....))).)))))).))))..)))...)))))........................... ( -18.60) >DroEre_CAF1 11674 108 + 1 UA---AUUAGUCCAUUAAUG-------UCCACUACAGGCCUAUCCCGAGUGGGACAUUUUCCAGACCUUCCACGCUGGAUCCUACCAGAGCGCUCUGGUCAACGAUGCCAGCGCCAAU ..---....(((....((((-------((((((...((......)).))).))))))).....)))......(((((((((..((((((....))))))....))).))))))..... ( -35.00) >DroYak_CAF1 14377 103 + 1 UA---AUUUG-----UAAUU-------UCGAUUACAGGCCUAUCCCGAGUGGGAUAUUUUCCAAACCUUCCAUGCUGGUUCCUACCAGAGCGCUCUGGUCAAUGAUGCCAGCGCCAAU ..---...((-----((((.-------...))))))(((.((((((....)))))).................((((((..(.((((((....))))))....)..)))))))))... ( -33.70) >DroAna_CAF1 18479 103 + 1 UAUUCGUUUG-----UACGU-------U---UCGUAGGCCUAUCCCGAGUGGGACAUCUUCCAGACCUUCCAUGCCGGCUCCUACCAAAGCGCUCUGGUAUACGAUGCCAGUGCCAGU ..........-----...((-------(---(.(((((......(((.((((((..((.....))..))))))..)))..)))))..))))((.(((((((...))))))).)).... ( -28.50) >consensus UA___AUUUG_____UAAUU_______UCGAUUACAGGCCUAUCCCGAGUGGGACAUUUUCCAGACCUUCCAUGCUGGCUCCUACCAGAGCGCUCUGGUCAACGAUGCCAGCGCCAAU ....................................(((.........((((((..((.....))..))))))((((((((..((((((....))))))....)).)))))))))... (-18.13 = -19.13 + 1.00)

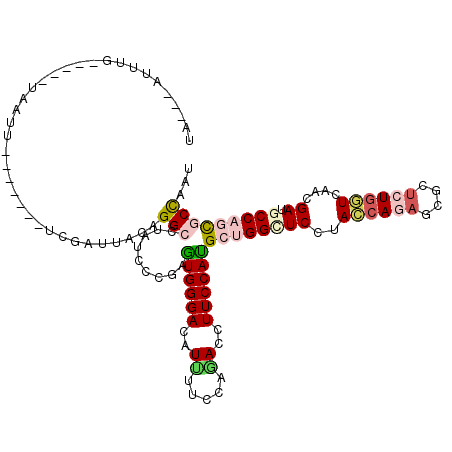

| Location | 17,577,128 – 17,577,231 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.55 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -20.35 |
| Energy contribution | -19.77 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17577128 103 - 27905053 AUUAGCGCUGGCAUCAUUCACCAGAGCGCUCUGGUACGAUCCAGCGUGGAAGGUCUGGAAAAUGUCCCACUCGGGAUAGGCCUGUAAUCGA-------AAUUA-----CAAAU---UA ....(((((((.(((....((((((....))))))..))))))))))...((((((.......(((((....)))))))))))(((((...-------.))))-----)....---.. ( -36.91) >DroVir_CAF1 15391 107 - 1 AUUGGGGUUGGCAUCAGCAAUGAGAGCGCUCUGAUAAGAGCCAGCAUGGAAGGUUUGGAAUAUAUCCCAUUCUGGAUAGGCCUUGGACAGA------AAAUCAAUU--GAAGU---UA (((((.((((((.(((....)))..))(((((....)))))))))..((...(((..((((.......))))..)))...)).........------...))))).--.....---.. ( -25.80) >DroPse_CAF1 12309 110 - 1 AGUGGGAGUGGCGUCGUUGAUCAGGGCGCUUUGGUAGGAGCCGGCAUGGAAGAUCUGGAAAAUGUCCCAUUCCGGAUAGGCCUGCAAUAGAUUGGUAUAAUUA-----UAAAG---CU .........(((....(..(((..(((.(((....))).))).(((.((...((((((((.........))))))))...)))))....)))..)(((....)-----))..)---)) ( -26.50) >DroEre_CAF1 11674 108 - 1 AUUGGCGCUGGCAUCGUUGACCAGAGCGCUCUGGUAGGAUCCAGCGUGGAAGGUCUGGAAAAUGUCCCACUCGGGAUAGGCCUGUAGUGGA-------CAUUAAUGGACUAAU---UA .((.(((((((.(((....((((((....))))))..)))))))))).)).((((((...(((((((.(((((((.....)))).))))))-------))))..))))))...---.. ( -47.40) >DroYak_CAF1 14377 103 - 1 AUUGGCGCUGGCAUCAUUGACCAGAGCGCUCUGGUAGGAACCAGCAUGGAAGGUUUGGAAAAUAUCCCACUCGGGAUAGGCCUGUAAUCGA-------AAUUA-----CAAAU---UA ...((((((((..((....((((((....))))))..)).))))).................((((((....)))))).)))((((((...-------.))))-----))...---.. ( -34.10) >DroAna_CAF1 18479 103 - 1 ACUGGCACUGGCAUCGUAUACCAGAGCGCUUUGGUAGGAGCCGGCAUGGAAGGUCUGGAAGAUGUCCCACUCGGGAUAGGCCUACGA---A-------ACGUA-----CAAACGAAUA .(((...(((((.((...(((((((....))))))).)))))))..))).((((((.......(((((....)))))))))))....---.-------.(((.-----...))).... ( -30.31) >consensus AUUGGCGCUGGCAUCAUUGACCAGAGCGCUCUGGUAGGAGCCAGCAUGGAAGGUCUGGAAAAUGUCCCACUCGGGAUAGGCCUGUAAUAGA_______AAUUA_____CAAAU___UA .........(((........(((((.(.((((((......))))...))..).)))))....((((((....)))))).))).................................... (-20.35 = -19.77 + -0.58)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:34 2006