
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,576,478 – 17,576,605 |
| Length | 127 |
| Max. P | 0.998725 |

| Location | 17,576,478 – 17,576,574 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.84 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -4.03 |
| Structure conservation index | 0.70 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17576478 96 + 27905053 UUAAAAUCUUUAUAAUUAUUAAAGUAUGCCACUUUGUCUUUCUCGCCAUAAUUUAAUGACAAAGAUACGGUGUAAUCAGUGCUU-UACAAGCACAAU .......(((((.......)))))((((((.(((((((...................)))))))....))))))....(((((.-....)))))... ( -18.31) >DroSec_CAF1 11722 96 + 1 UUAAAAUCUUUAUAAUUAUAAAAGUAUGCCACUGUGUCUUUGUCGUCAUAAUUGUAUGACAAAGAUACGGUGUUAUCAGUGCUU-UACAAUCACAAG ....................(((((((..(((((((((((((((((((....)).)))))))))))))))))......))))))-)........... ( -29.80) >DroSim_CAF1 10929 96 + 1 UUAAAAUCUUUAUAAUUAUAAAAGUAUGCCACUGUGUCUUUGUCGUCAUAAUUUUAUGACAAAGAUACGGUGUUAUCAGUGCUU-UACAAGCACAAG .......................(.(((.(((((((((((((((((.(.....).))))))))))))))))).)))).(((((.-....)))))... ( -29.90) >DroEre_CAF1 11035 85 + 1 UUAAAAUGUUUAUACUCAUUAAAGUAUGCCACCGUGUCUUUGUCGUCAUGGGUUUAUGACGAAGAUACGGUGAUAUCA------------GCACAAG ......((((((((((......)))))).(((((((((((((((((.(.....).))))))))))))))))).....)------------))).... ( -28.80) >DroYak_CAF1 13723 97 + 1 UUAAAAUGUUUAUAACCACUUAAGUAUGUCACUGCCUCUUUGUCGUCAUGGGUUUAUGACAAAGAUACGGCGAUAUCAGUGGUUCUACAAGCACAAC ......(((((..(((((((...(.(((((.(((..((((((((((.(.....).))))))))))..))).)))))))))))))....))))).... ( -27.50) >consensus UUAAAAUCUUUAUAAUUAUUAAAGUAUGCCACUGUGUCUUUGUCGUCAUAAUUUUAUGACAAAGAUACGGUGUUAUCAGUGCUU_UACAAGCACAAG .......................(.(((.(((((((((((((((((.........))))))))))))))))).)))).................... (-18.76 = -19.84 + 1.08)



| Location | 17,576,478 – 17,576,574 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -12.02 |
| Energy contribution | -11.58 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17576478 96 - 27905053 AUUGUGCUUGUA-AAGCACUGAUUACACCGUAUCUUUGUCAUUAAAUUAUGGCGAGAAAGACAAAGUGGCAUACUUUAAUAAUUAUAAAGAUUUUAA ...(((((....-.)))))......(((..(.((((((((((......)))))))...))).)..))).....(((((.......)))))....... ( -16.20) >DroSec_CAF1 11722 96 - 1 CUUGUGAUUGUA-AAGCACUGAUAACACCGUAUCUUUGUCAUACAAUUAUGACGACAAAGACACAGUGGCAUACUUUUAUAAUUAUAAAGAUUUUAA .(((((((((((-(((...((....(((.((.((((((((.............)))))))).)).))).))...))))))))))))))......... ( -23.82) >DroSim_CAF1 10929 96 - 1 CUUGUGCUUGUA-AAGCACUGAUAACACCGUAUCUUUGUCAUAAAAUUAUGACGACAAAGACACAGUGGCAUACUUUUAUAAUUAUAAAGAUUUUAA .(((((.(((((-(((...((....(((.((.((((((((.............)))))))).)).))).))...)))))))).)))))......... ( -19.72) >DroEre_CAF1 11035 85 - 1 CUUGUGC------------UGAUAUCACCGUAUCUUCGUCAUAAACCCAUGACGACAAAGACACGGUGGCAUACUUUAAUGAGUAUAAACAUUUUAA .((((((------------(....(((((((.((((((((((......)))))))...))).)))))))(((......))))))))))......... ( -25.00) >DroYak_CAF1 13723 97 - 1 GUUGUGCUUGUAGAACCACUGAUAUCGCCGUAUCUUUGUCAUAAACCCAUGACGACAAAGAGGCAGUGACAUACUUAAGUGGUUAUAAACAUUUUAA ...(((.(((((..((((((.((.((((.((.((((((((.............)))))))).)).)))).)).....))))))))))).)))..... ( -25.52) >consensus CUUGUGCUUGUA_AAGCACUGAUAACACCGUAUCUUUGUCAUAAAAUUAUGACGACAAAGACACAGUGGCAUACUUUAAUAAUUAUAAAGAUUUUAA .........................(((.((.((((((((((......)))))))...))).)).)))............................. (-12.02 = -11.58 + -0.44)

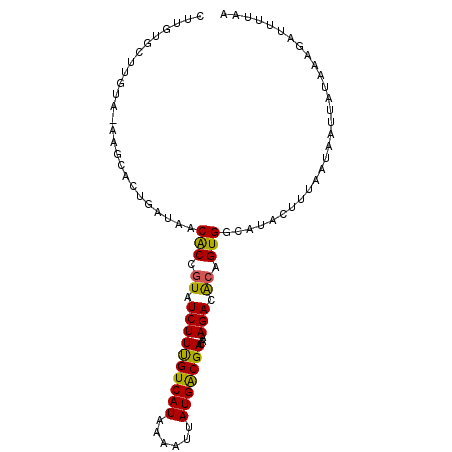

| Location | 17,576,509 – 17,576,605 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -14.98 |
| Energy contribution | -15.66 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17576509 96 + 27905053 CUUUGUCUUUCUCGCCAUAAUUUAAUGACAAAGAUACGGUGUAAUCAGUGCUU-UACAAGCACAAUGUCUUUAAUGGAUGCAUAAAUUAAUUGUUAU (((((((...................))))))).....((((.....(((((.-....)))))...((((.....)))))))).............. ( -16.81) >DroSec_CAF1 11753 96 + 1 CUGUGUCUUUGUCGUCAUAAUUGUAUGACAAAGAUACGGUGUUAUCAGUGCUU-UACAAUCACAAGGUCUUUAGUGGAUGCAUAAAUGAAUUGUUAU (((((((((((((((((....)).)))))))))))))))..((((..((((((-(((................))))).))))..))))........ ( -26.99) >DroSim_CAF1 10960 96 + 1 CUGUGUCUUUGUCGUCAUAAUUUUAUGACAAAGAUACGGUGUUAUCAGUGCUU-UACAAGCACAAGGUCUUUAGUGGAUGCAUAAAUGAAUUGUUAU (((((((((((((((.(.....).))))))))))))))).....((((((((.-....)))))...((((.....)))).......)))........ ( -27.50) >DroEre_CAF1 11066 85 + 1 CCGUGUCUUUGUCGUCAUGGGUUUAUGACGAAGAUACGGUGAUAUCA------------GCACAAGCGCUUGAAUGGAUGCAUAAAUUAAUUGUUAU (((((((((((((((.(.....).)))))))))))))))((((((((------------((......)).)))(((....)))........))))). ( -23.30) >DroYak_CAF1 13754 97 + 1 CUGCCUCUUUGUCGUCAUGGGUUUAUGACAAAGAUACGGCGAUAUCAGUGGUUCUACAAGCACAACCGCUUGAAUGAAUGCAUAAUUUAAUUGCCAU .....((((((((((.(.....).))))))))))...((((((....(((((((..(((((......)))))...)))).)))......)))))).. ( -26.30) >consensus CUGUGUCUUUGUCGUCAUAAUUUUAUGACAAAGAUACGGUGUUAUCAGUGCUU_UACAAGCACAAGGUCUUUAAUGGAUGCAUAAAUUAAUUGUUAU (((((((((((((((.........))))))))))))))).......................................................... (-14.98 = -15.66 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:32 2006