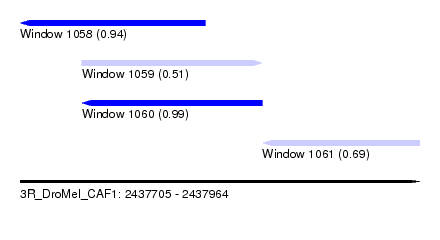
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,437,705 – 2,437,964 |
| Length | 259 |
| Max. P | 0.994585 |

| Location | 2,437,705 – 2,437,825 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -45.04 |
| Consensus MFE | -34.30 |
| Energy contribution | -36.30 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
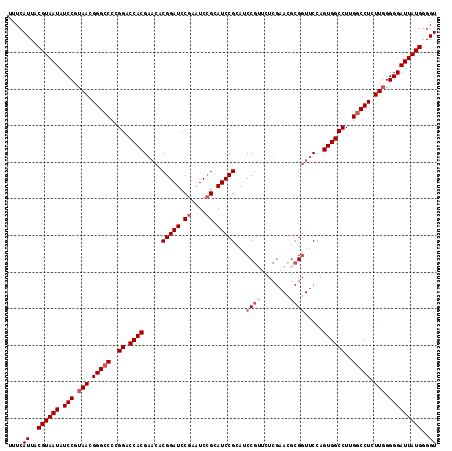
>3R_DroMel_CAF1 2437705 120 - 27905053 CGCGUUCGAGAACGGAUGCGGAUGCUGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCCCCCGUGCGUGGGCUUCAAUUAAAUU (((((.(((..((((((((((((.(......).))))))))))))..)).(((((.(((((((.......)))))))...((........))))))).).)))))............... ( -43.80) >DroSec_CAF1 4069 119 - 1 CGCGUUCGAGAACGGAUGCGGAUGCGGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCC-CCGUGCGUGGGCUUCAAUUAAAUU (((((..((..((((((((((((.((....)).))))))))))))..))((((((.(((((((.......)))))))...((........)))))-))).)))))............... ( -46.90) >DroSim_CAF1 4164 120 - 1 CGCGUUCGAGAACGGAUGCGGAUGCGGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCCCCCGUGCGUGGGCUUCAAUUAAAUU (((((.((.....((((((((((((((((....)))))))))))).))))(((((.(((((((.......)))))))...((........))))))))).)))))............... ( -46.30) >DroEre_CAF1 4152 120 - 1 CGCAUUCGAGUCCGGCUCCGGAUCCGGACUCGGAUCCGCGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCCCCCGUGCGUGGGCUUCAAUUAAAUU ...(((.((((((((((((((((((((((.((....)).))))).)))))))))))(((((((.......))))))).............((((......)))))))))).)))...... ( -51.20) >DroYak_CAF1 3767 111 - 1 C---------ACUGGCUCCGGAUUCGGAUUCGGAUCCGUGUUCGUGGUCCGUGCCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCCCCCGUGCGUGGGCUUCAAUUAAAUU .---------((.(((..(((((.(((((....))))).)))))..))).))(((((((.((((......((((((...........))))))...))))))).))))............ ( -37.00) >consensus CGCGUUCGAGAACGGAUGCGGAUGCGGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCCCCCGUGCGUGGGCUUCAAUUAAAUU .............((((((((((((((((....)))))))))))).))))(((((((((((((.......))))))..............((((......)))))))))))......... (-34.30 = -36.30 + 2.00)

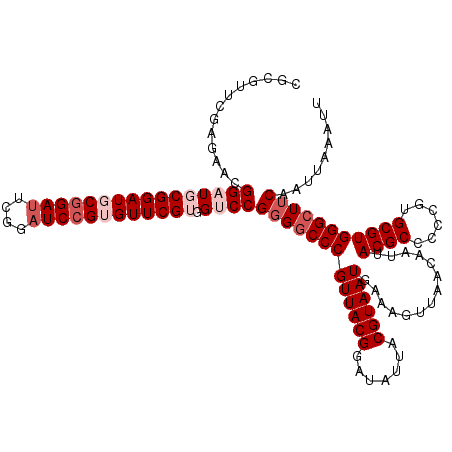
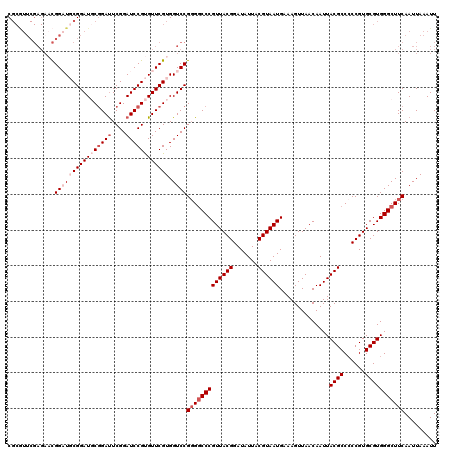
| Location | 2,437,745 – 2,437,862 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -43.22 |
| Consensus MFE | -34.54 |
| Energy contribution | -35.94 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2437745 117 + 27905053 UUUCAUUACGUAAUAUCCGUAACGGGCCCCGGACCACGAACACGGAUCCGAAUCAGCAUCCGCAUCCGUUCUCGAACGCGGUUCCAGUGGCCUUGGCCUCUUGGGGGAUUAUGGGGU ....((..((((((.(((.(((.(((((..((.((((((..((((((.((..........)).))))))..))((((...))))..))))))..))))).))).)))))))))..)) ( -40.60) >DroSec_CAF1 4108 117 + 1 UUUCAUUACGUAAUAUCCGUAACGGGCCCCGGACCACGAACACGGAUCCGAAUCCGCAUCCGCAUCCGUUCUCGAACGCGGUUCCAGUGGCCUUGGCCUCUUGGGGGAUUAUGGGGU ....((..((((((.(((.(((.(((((..((.((((((((.(((((.((....)).)))))...(((((....)))).)))))..))))))..))))).))).)))))))))..)) ( -42.00) >DroSim_CAF1 4204 117 + 1 UUUCAUUACGUAAUAUCCGUAACGGGCCCCGGACCACGAACACGGAUCCGAAUCCGCAUCCGCAUCCGUUCUCGAACGCGGUUCCAGUGGCCUUGGCCUCUUGGGGGAUUAUGGGGU ....((..((((((.(((.(((.(((((..((.((((((((.(((((.((....)).)))))...(((((....)))).)))))..))))))..))))).))).)))))))))..)) ( -42.00) >DroEre_CAF1 4192 117 + 1 UUUCAUUACGUAAUAUCCGUAACGGGCCCCGGACCACGAACGCGGAUCCGAGUCCGGAUCCGGAGCCGGACUCGAAUGCGGUUCGAGUGGCCUUGGCCUCUUUGGGGAUUAUGGGGU ....((..((((((..(((.((.(((((..((.((((((((.((.((.(((((((((........))))))))).)).))))))..))))))..))))).)))))..))))))..)) ( -52.50) >DroYak_CAF1 3807 106 + 1 UUUCAUUACGUAAUAUCCGUAACGGGGCACGGACCACGAACACGGAUCCGAAUCCGAAUCCGGAGCCAGU---------G--GCCAGUGGCCUUGGCCUCUUGGGGGAUUAUGGGGU ....((..((((((.(((.(((.(((((..((.((((...(((((.((((..........)))).)).))---------)--....))))))...)))))))).)))))))))..)) ( -39.00) >consensus UUUCAUUACGUAAUAUCCGUAACGGGCCCCGGACCACGAACACGGAUCCGAAUCCGCAUCCGCAUCCGUUCUCGAACGCGGUUCCAGUGGCCUUGGCCUCUUGGGGGAUUAUGGGGU ....((..((((((.(((.(((.(((((..((.((((.....(((((.((....)).)))))...((((........)))).....))))))..))))).))).)))))))))..)) (-34.54 = -35.94 + 1.40)

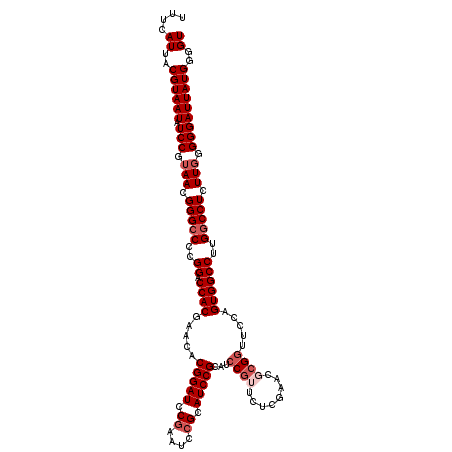
| Location | 2,437,745 – 2,437,862 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -48.00 |
| Consensus MFE | -37.58 |
| Energy contribution | -40.46 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
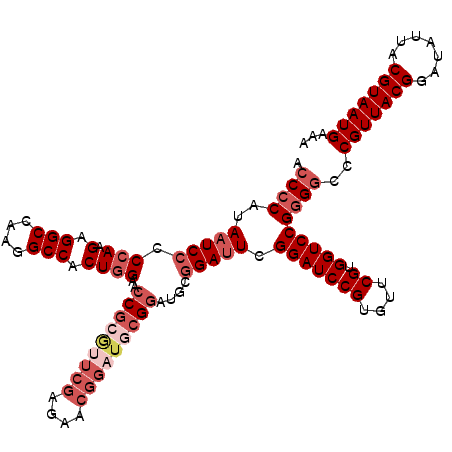
>3R_DroMel_CAF1 2437745 117 - 27905053 ACCCCAUAAUCCCCCAAGAGGCCAAGGCCACUGGAACCGCGUUCGAGAACGGAUGCGGAUGCUGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAA .((((...........((.(((....))).))(((.(((((.......((((((.(((((....))))).))))))...))))))))))))..(((((((.......)))))))... ( -47.70) >DroSec_CAF1 4108 117 - 1 ACCCCAUAAUCCCCCAAGAGGCCAAGGCCACUGGAACCGCGUUCGAGAACGGAUGCGGAUGCGGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAA .((((..(((((.(((.(.(((....))).))))..(((((((((....)))))))))....))))).(((((((....)).)))))))))..(((((((.......)))))))... ( -51.00) >DroSim_CAF1 4204 117 - 1 ACCCCAUAAUCCCCCAAGAGGCCAAGGCCACUGGAACCGCGUUCGAGAACGGAUGCGGAUGCGGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAA .((((..(((((.(((.(.(((....))).))))..(((((((((....)))))))))....))))).(((((((....)).)))))))))..(((((((.......)))))))... ( -51.00) >DroEre_CAF1 4192 117 - 1 ACCCCAUAAUCCCCAAAGAGGCCAAGGCCACUCGAACCGCAUUCGAGUCCGGCUCCGGAUCCGGACUCGGAUCCGCGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAA ...................(((....)))(((((((.....)))))))..((((((((((((((((.((....)).))))).)))))))))))(((((((.......)))))))... ( -52.50) >DroYak_CAF1 3807 106 - 1 ACCCCAUAAUCCCCCAAGAGGCCAAGGCCACUGGC--C---------ACUGGCUCCGGAUUCGGAUUCGGAUCCGUGUUCGUGGUCCGUGCCCCGUUACGGAUAUUACGUAAUGAAA ...................(((....)))((.(((--(---------((..((..((((((((....)))))))).))..)))))).))....(((((((.......)))))))... ( -37.80) >consensus ACCCCAUAAUCCCCCAAGAGGCCAAGGCCACUGGAACCGCGUUCGAGAACGGAUGCGGAUGCGGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAA .((((..(((((.(((.(.(((....))).))))..(((((((((....)))))))))....))))).(((((((....)).)))))))))..(((((((.......)))))))... (-37.58 = -40.46 + 2.88)

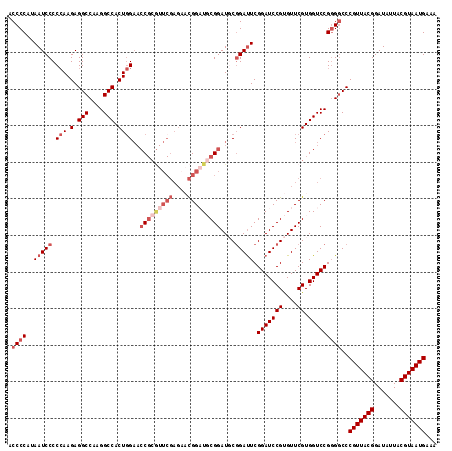
| Location | 2,437,862 – 2,437,964 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
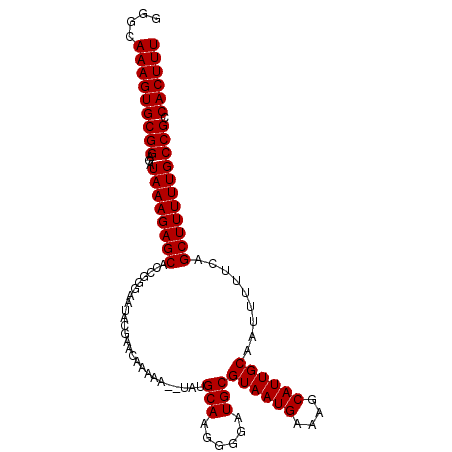
>3R_DroMel_CAF1 2437862 102 - 27905053 GGGCAAAGUGCGGAGAUAAAGAGCACCGUGAAUACGAACAAAAA--UAUGCAAGGGGAUGCGUAAUGAAAGCAUUGCAAUUUUUCAGCUUUUUGCCGCCACUUU ....(((((((((...((((((((..(((....)))........--.......(..((...((((((....))))))...))..).))))))))))).)))))) ( -25.70) >DroSec_CAF1 4225 102 - 1 GGGCAAAGUGCGGAGAUAAAGAGCACCGGGAAUACGAACAAAAA--UAUGCAAGGGGAUGCGUAAUGAAAGCAUUGCAAUUUUUCAGCUUUUUGCCGCCACUUU ....(((((((((...((((((((..((......))........--.......(..((...((((((....))))))...))..).))))))))))).)))))) ( -25.90) >DroSim_CAF1 4321 102 - 1 GGGCAAAGUGCGGAGAUAAAGAGCACCGGGAAUACGAACAAAAA--UAUGCAAGGGGAUGCGUAAUGAAAGCAUUGCAAUUUUUCAGCUUUUUGCCGCCACUUU ....(((((((((...((((((((..((......))........--.......(..((...((((((....))))))...))..).))))))))))).)))))) ( -25.90) >DroEre_CAF1 4309 102 - 1 GGGCAAAGUGCGGAGAUAAAGAGCACCGGGAAUACCGACAAAAA--UAUGCAAUGGGAUGCGUAAUGAAAGCAUUGCAAUUUUUCAGCUUUUUGCCGCCACUUU ....(((((((((...((((((((..(((.....)))..(((((--(..(((......)))((((((....)))))).))))))..))))))))))).)))))) ( -28.90) >DroYak_CAF1 3913 104 - 1 GGGCAAAGUGCGGAGAUAAAGAGCACCGGGAAUAUCGACAACAAUAUAUGCAAUGGGAUGCGUAAUGAAAGCAUUGCAAUUUUUCAGCUUUUUGCCGCCACUUU ....(((((((((...((((((((..(((.....)))................((.(((((.........))))).))........))))))))))).)))))) ( -25.10) >consensus GGGCAAAGUGCGGAGAUAAAGAGCACCGGGAAUACGAACAAAAA__UAUGCAAGGGGAUGCGUAAUGAAAGCAUUGCAAUUUUUCAGCUUUUUGCCGCCACUUU ....(((((((((...((((((((.........................(((......)))((((((....)))))).........))))))))))).)))))) (-24.50 = -24.50 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:53 2006