| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,560,919 – 17,561,062 |
| Length | 143 |
| Max. P | 0.999986 |

| Location | 17,560,919 – 17,561,024 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -20.38 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17560919 105 - 27905053 CAAAUCCACUAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGUUGCAUCGAAUAUUUAUAUAAGCAAUUUCUU--------UCGG-------UUUUUGCAUUUGAAAUUC ((((...((((((((((....)))....(((((((....))))))))))))))(((((((((....................)--------))))-------....))))))))...... ( -17.45) >DroSec_CAF1 80 113 - 1 CAAAGCAGCCAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGCUGCAUCGAAUAUUUAUACAAGGAAUUUCUUCGGCUUUUUCCA-------AUUUUGCAUUUGAAAUUC ....(((((((((((((....)))....(((((((....)))))))))))))))))..................(((((((....((........-------.....))....))))))) ( -32.12) >DroSim_CAF1 80 113 - 1 CAAAGCCGCCAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGCUGCAUCGAAUAUUUAUACAAGGAAUUUCUUCGGGUUUUUCGA-------UUUUUGCAUUUGAAACUC ....((.((((((((((....)))....(((((((....)))))))))))))).)).((((((..........((((..((....))..))))..-------.......))))))..... ( -29.01) >DroEre_CAF1 1369 120 - 1 CAAAGCCGCCAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGCUGCAUCGAAUAUUUAUACAAGGAAUUUCUUUUGGUUUUUCGUUUCUUGCUUUUUGCAUUUGAAAUCC ....((.((((((((((....)))....(((((((....)))))))))))))).)).................((((..((....))..))))(((((.(((.....)))...))))).. ( -29.10) >DroYak_CAF1 1291 113 - 1 CAAAGCCGCCAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGCUGCAUCGAAUAUUUAUACAAGGAAUUUCUUCGGGUUUUUCGG-------UUUUUGCAUUUGAAAUUC ....((.((((((((((....)))....(((((((....)))))))))))))).)).((((((..........((((..((....))..)))).(-------(....))))))))..... ( -29.50) >consensus CAAAGCCGCCAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGCUGCAUCGAAUAUUUAUACAAGGAAUUUCUUCGGGUUUUUCGA_______UUUUUGCAUUUGAAAUUC ((((((.((((((((((....)))....(((((((....)))))))))))))).)).................((((..((....))..)))).................))))...... (-20.38 = -21.30 + 0.92)


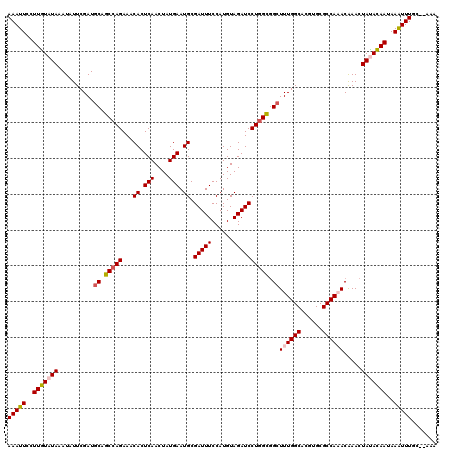
| Location | 17,560,944 – 17,561,062 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17560944 118 + 27905053 AAAUUGCUUAUAUAAAUAUUCGAUGCAACCAGAAACACUCAACUAUGAAUGCGAUUUCCAUGUAGAUCCUAGUGGAUUUGGCACGUGCGCCAAACGAACUAUAUAAUAAAUUUGC--AAA .......................(((((...((((((.(((....))).))...)))).((((((.(((....)))((((((......))))))....)))))).......))))--).. ( -19.70) >DroSec_CAF1 113 120 + 1 AAAUUCCUUGUAUAAAUAUUCGAUGCAGCCAGAAACACUCAACUAUGAAUGCGAUUUCCAUGUAGAUCCUGGCUGCUUUGGCACGUGCGCCAAACAAACUAUACAAUAAAUUUGCGAAAA (((((..(((((((..........((((((((...((.(((....))).)).(((((......)))))))))))))((((((......)))))).....)))))))..)))))....... ( -31.60) >DroSim_CAF1 113 120 + 1 AAAUUCCUUGUAUAAAUAUUCGAUGCAGCCAGAAACACUCAACUAUGAAUGCGAUUUCCAUGUAGAUCCUGGCGGCUUUGGCACGUGCGCCAAACAAACUAUACAAUAAAUUUGCGAAAA (((((..(((((((..........((.(((((...((.(((....))).)).(((((......)))))))))).))((((((......)))))).....)))))))..)))))....... ( -27.00) >DroEre_CAF1 1409 117 + 1 AAAUUCCUUGUAUAAAUAUUCGAUGCAGCCAGAAACACUCAACUAUGAAUGCGAUUUCCAUGUAGAUCCUGGCGGCUUUGGCACGUGCGCCA-ACAAACUACACAAUAAGUUUGC--AAA ........................((.(((((...((.(((....))).)).(((((......)))))))))).)).(((((......))))-)((((((........)))))).--... ( -23.50) >DroYak_CAF1 1324 117 + 1 AAAUUCCUUGUAUAAAUAUUCGAUGCAGCCAGAAACACUCAACUAUGAAUGCGAUUUCCAUGUAGAUCCUGGCGGCUUUGGCACGUGCGCCA-ACAAACUACACAAUAAGUUUGC--AAA ........................((.(((((...((.(((....))).)).(((((......)))))))))).)).(((((......))))-)((((((........)))))).--... ( -23.50) >consensus AAAUUCCUUGUAUAAAUAUUCGAUGCAGCCAGAAACACUCAACUAUGAAUGCGAUUUCCAUGUAGAUCCUGGCGGCUUUGGCACGUGCGCCAAACAAACUAUACAAUAAAUUUGC__AAA (((((..(((((((..........((.(((((...((.(((....))).)).(((((......)))))))))).))((((((......)))))).....)))))))..)))))....... (-22.10 = -22.42 + 0.32)

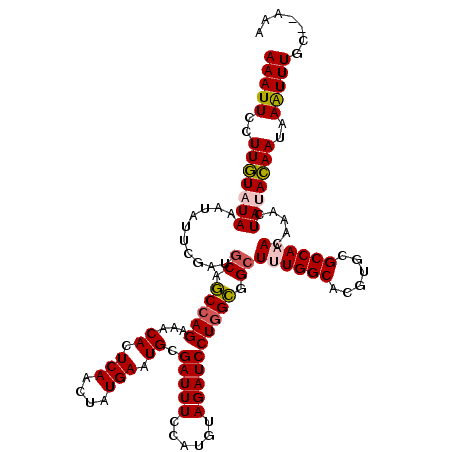
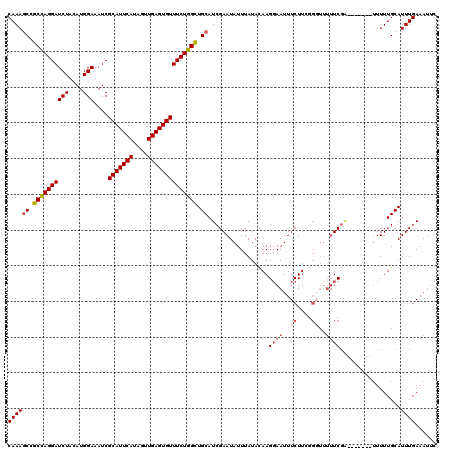
| Location | 17,560,944 – 17,561,062 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -34.42 |
| Energy contribution | -34.42 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.96 |
| SVM decision value | 5.42 |
| SVM RNA-class probability | 0.999986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17560944 118 - 27905053 UUU--GCAAAUUUAUUAUAUAGUUCGUUUGGCGCACGUGCCAAAUCCACUAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGUUGCAUCGAAUAUUUAUAUAAGCAAUUU .((--((.......(((((((((((((((((((....)))))))...((((((((((....)))....(((((((....)))))))))))))).....)))))...)))))))))))... ( -28.11) >DroSec_CAF1 113 120 - 1 UUUUCGCAAAUUUAUUGUAUAGUUUGUUUGGCGCACGUGCCAAAGCAGCCAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGCUGCAUCGAAUAUUUAUACAAGGAAUUU .......((((((.(((((((((((((((((((....)))))))(((((((((((((....)))....(((((((....)))))))))))))))))..)))))...))))))).)))))) ( -42.80) >DroSim_CAF1 113 120 - 1 UUUUCGCAAAUUUAUUGUAUAGUUUGUUUGGCGCACGUGCCAAAGCCGCCAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGCUGCAUCGAAUAUUUAUACAAGGAAUUU .......((((((.(((((((((((((((((((....)))))))((.((((((((((....)))....(((((((....)))))))))))))).))..)))))...))))))).)))))) ( -38.20) >DroEre_CAF1 1409 117 - 1 UUU--GCAAACUUAUUGUGUAGUUUGU-UGGCGCACGUGCCAAAGCCGCCAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGCUGCAUCGAAUAUUUAUACAAGGAAUUU ...--.........(((((((((((((-(((((....)))))).((.((((((((((....)))....(((((((....)))))))))))))).))..)))))...)))))))....... ( -34.70) >DroYak_CAF1 1324 117 - 1 UUU--GCAAACUUAUUGUGUAGUUUGU-UGGCGCACGUGCCAAAGCCGCCAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGCUGCAUCGAAUAUUUAUACAAGGAAUUU ...--.........(((((((((((((-(((((....)))))).((.((((((((((....)))....(((((((....)))))))))))))).))..)))))...)))))))....... ( -34.70) >consensus UUU__GCAAAUUUAUUGUAUAGUUUGUUUGGCGCACGUGCCAAAGCCGCCAGGAUCUACAUGGAAAUCGCAUUCAUAGUUGAGUGUUUCUGGCUGCAUCGAAUAUUUAUACAAGGAAUUU .......((((((.(((((((((((((((((((....)))))))((.((((((((((....)))....(((((((....)))))))))))))).))..)))))...))))))).)))))) (-34.42 = -34.42 + 0.00)

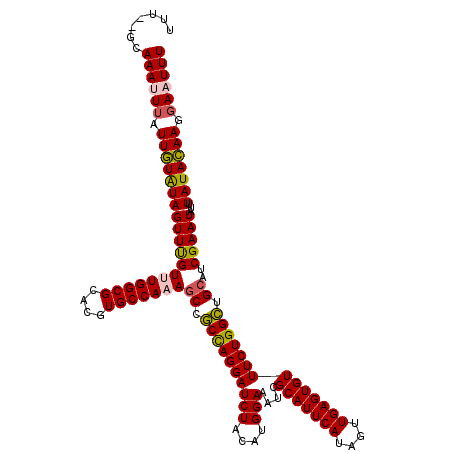
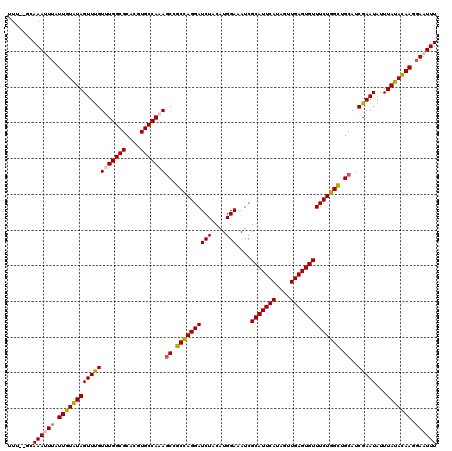
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:22 2006