
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,555,595 – 17,555,738 |
| Length | 143 |
| Max. P | 0.856223 |

| Location | 17,555,595 – 17,555,705 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -32.59 |
| Energy contribution | -32.70 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17555595 110 + 27905053 GAGCUUGAUGGACCCCUCUCAUCAAUUGAGAUAUUAUGAAAUCCACUGCACUCC--CCGCUUCCCCC-GCGAGUGCGUGCGGUCCGCGCUCGCUAAUAGAGACGCAUUCAUGU------- ..((.(((.(((((..(((((.....)))))............(((.((((((.--..((.......-))))))))))).)))))((((((.......))).)))..))).))------- ( -35.20) >DroSim_CAF1 3569 113 + 1 GAGCUUGAUGGACCCCUCUCAUCAAUUGAGAUAUUAUGAAAUCCACUGCACUCCCCCCGCUUCCCCCAGCGAGUGCGUGCGGUCCGCGCUCGCUAAUAGAGACGCAUUCAUGU------- ..((.(((.(((((..(((((.....)))))............(((.((((((.....(((......)))))))))))).)))))((((((.......))).)))..))).))------- ( -35.70) >DroEre_CAF1 7867 117 + 1 GAGCUUGAUGGACCCCUCUCAUCAAUUGAGAUAUUAUGAAAUCCACUGCACGCC--GCGCUUCCCCA-GCGAGUGCGUGCGGUCCGCGUUCGCUAAUAGAGACGCAUUCAUGUACAUUCA (.((.(((.(((((..(((((.....)))))................(((((((--.((((.....)-))).).)))))))))))((((((.......)).))))..))).)).)..... ( -39.30) >consensus GAGCUUGAUGGACCCCUCUCAUCAAUUGAGAUAUUAUGAAAUCCACUGCACUCC__CCGCUUCCCCC_GCGAGUGCGUGCGGUCCGCGCUCGCUAAUAGAGACGCAUUCAUGU_______ ..((.(((.(((((..(((((.....)))))............(((.(((((.....(((........))))))))))).)))))((((((.......))).)))..))).))....... (-32.59 = -32.70 + 0.11)

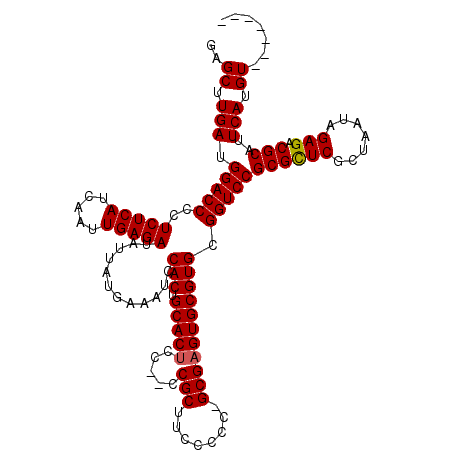
| Location | 17,555,595 – 17,555,705 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -36.57 |
| Energy contribution | -37.23 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17555595 110 - 27905053 -------ACAUGAAUGCGUCUCUAUUAGCGAGCGCGGACCGCACGCACUCGC-GGGGGAAGCGG--GGAGUGCAGUGGAUUUCAUAAUAUCUCAAUUGAUGAGAGGGGUCCAUCAAGCUC -------.......(((.(((((....(((((.(((.......))).)))))-.))))).))).--.((((...((((((..(......(((((.....))))))..))))))...)))) ( -41.20) >DroSim_CAF1 3569 113 - 1 -------ACAUGAAUGCGUCUCUAUUAGCGAGCGCGGACCGCACGCACUCGCUGGGGGAAGCGGGGGGAGUGCAGUGGAUUUCAUAAUAUCUCAAUUGAUGAGAGGGGUCCAUCAAGCUC -------.......(((.(((((..(((((((.(((.......))).)))))))))))).)))....((((...((((((..(......(((((.....))))))..))))))...)))) ( -42.30) >DroEre_CAF1 7867 117 - 1 UGAAUGUACAUGAAUGCGUCUCUAUUAGCGAACGCGGACCGCACGCACUCGC-UGGGGAAGCGC--GGCGUGCAGUGGAUUUCAUAAUAUCUCAAUUGAUGAGAGGGGUCCAUCAAGCUC ....(((((.....((((..(((.(((((((..(((.......)))..))))-))))))..)))--)..)))))((((((..(......(((((.....))))))..))))))....... ( -39.40) >consensus _______ACAUGAAUGCGUCUCUAUUAGCGAGCGCGGACCGCACGCACUCGC_GGGGGAAGCGG__GGAGUGCAGUGGAUUUCAUAAUAUCUCAAUUGAUGAGAGGGGUCCAUCAAGCUC ..............(((.(((((....(((((.(((.......))).)))))..))))).)))....((((...((((((..(......(((((.....))))))..))))))...)))) (-36.57 = -37.23 + 0.67)



| Location | 17,555,635 – 17,555,738 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.26 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -33.05 |
| Energy contribution | -33.50 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17555635 103 + 27905053 AUCCACUGCACUCC--CCGCUUCCCCC-GCGAGUGCGUGCGGUCCGCGCUCGCUAAUAGAGACGCAUUCAUGU--------------ACAUCCUUCUGGACCUCGCGCUGAGCUCCACGG .......((.....--..)).....((-(.(((((((((.(((((((((((.......))).)))((....))--------------..........))))).)))))...))))..))) ( -30.60) >DroSim_CAF1 3609 106 + 1 AUCCACUGCACUCCCCCCGCUUCCCCCAGCGAGUGCGUGCGGUCCGCGCUCGCUAAUAGAGACGCAUUCAUGU--------------ACAUCCUUCUGGACCUCGCGCUGAGCUCCACGG ................(((((......)))(((((((((.(((((((((((.......))).)))((....))--------------..........))))).)))))...))))...)) ( -30.90) >DroEre_CAF1 7907 113 + 1 AUCCACUGCACGCC--GCGCUUCCCCA-GCGAGUGCGUGCGGUCCGCGUUCGCUAAUAGAGACGCAUUCAUGUACAUUCAUGUAUGUACAUCCCUCUGGACCUCGCGCUGAGCCC----G .......(((((((--.((((.....)-))).).))))))(((((((((((.......)).))))....((((((((......))))))))......)))))..((.....))..----. ( -41.30) >consensus AUCCACUGCACUCC__CCGCUUCCCCC_GCGAGUGCGUGCGGUCCGCGCUCGCUAAUAGAGACGCAUUCAUGU______________ACAUCCUUCUGGACCUCGCGCUGAGCUCCACGG .......((........(((........)))...(((((.(((((((((((.......))).)))....((((((((......))))))))......))))).)))))...))....... (-33.05 = -33.50 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:17 2006