
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,479,149 – 17,479,252 |
| Length | 103 |
| Max. P | 0.545460 |

| Location | 17,479,149 – 17,479,252 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.13 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -9.75 |
| Energy contribution | -9.20 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17479149 103 - 27905053 UUGGUGUUCUUUUCGCCAGUUUCGAUCUUC----UCUGAGGAUCGCGUUUCAAUACUCUUUGAAUGGAAUAAAUCUGGUUUGGUGG----------AAGAACUUCUGAAAAUUCGGA .....(((((((.((((((...((((((((----...))))))))((((.(((......))))))).............)))))))----------)))))).(((((....))))) ( -27.70) >DroSec_CAF1 31211 106 - 1 UUGGUGUUCUUUGCGCCGGUUUCGAUCUUCUUACUCGGAGGAUCGCGU-UCGAUACUCUUUGAAUGGAAUAAAUCUAGUUUGGAGG----------AAGAACUUCUGAAAAUUCUGA ..(((((.....)))))((((((((((((((.....)))))))))(((-((((......)))))))....)))))...((..((((----------.....))))..))........ ( -30.00) >DroSim_CAF1 32216 106 - 1 UUGGUGUUCUUUGCGCCGGUUUCGAUCUUCUUGCUCUGAGGAUCGCGU-UCGAUACUCUUUGAAUGGAAUAAAUCUAGUUUGGUGU----------AAGAACUUCUGAAAAUUCGGA .....((((((.(((((((...((((((((.......))))))))(((-((((......))))))).............)))))))----------)))))).(((((....))))) ( -32.90) >DroEre_CAF1 31600 106 - 1 UUGGUGUUCUUUUUGCCACUGCCGAUCUUCUUUUUCUGAGGAUCUCGU-UGGAUGCUCUUCGAAUGGAAUAAAUCUGGUCCGGUGG----------AAGAACUUCUGAAUUUUUUAA (..(.((((((....(((((((((((((((.......)))))))((((-(.((......)).))))).........)))..)))))----------))))))..)..)......... ( -29.10) >DroYak_CAF1 32288 106 - 1 UUGGUGUUCUUUUUGCCGCUUGCGAUCUUUUUGCUCUGAGGAUAUCGU-UCGAUGCUCUUUGAAUGGAAUAAAUCCGGUCUGCUGC----------AAGAACUUCUGAAAUUUAUAA (..(.(((((...(((.((..((((.....)))).....((((.((((-((((......)))))))).....)))).....)).))----------))))))..)..)......... ( -23.20) >DroAna_CAF1 50586 115 - 1 UUAGUGUUUCUUCUGGCUCUCAGAUUCUUCUU--UCUGAUACUCUUUUUUUUUUUUUUUUUGAAUAAAGGAGAGUAGCUUUGGUAUUUCUUUUUAAAUAAGGCUCUGAAAUAUAUAC ...(((((((....((((.(((((........--)))))(((((((((((.(((.......))).)))))))))))........................))))..))))))).... ( -23.60) >consensus UUGGUGUUCUUUUCGCCGCUUUCGAUCUUCUU_CUCUGAGGAUCGCGU_UCGAUACUCUUUGAAUGGAAUAAAUCUGGUUUGGUGG__________AAGAACUUCUGAAAAUUCUAA ((((.((((((...((((....((((((((.......))))))))........((.(((......))).))....)))).................))))))..))))......... ( -9.75 = -9.20 + -0.55)

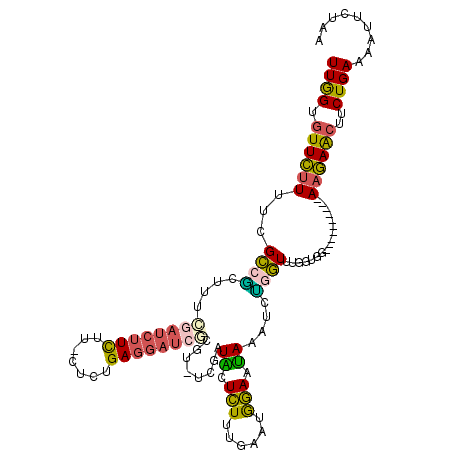

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:51 2006