| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,449,290 – 17,449,386 |
| Length | 96 |
| Max. P | 0.654675 |

| Location | 17,449,290 – 17,449,386 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.92 |
| Mean single sequence MFE | -23.81 |
| Consensus MFE | -15.48 |
| Energy contribution | -14.62 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
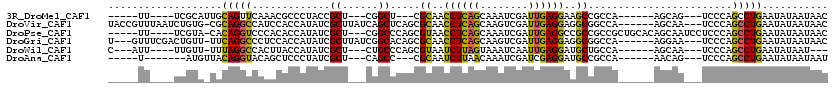
>3R_DroMel_CAF1 17449290 96 - 27905053 -----UU----UCGCAUUGCAGUUCAAACGCCCUACCGCU---CGGCU---CGCAACCUCAGCAAAUCGAUUGAGGAAGCCGCCA------AGCAG---UCCCAGCCUGAAUAUAAUAAC -----..----..........(((((...((...((.(((---.(((.---.((..(((((((.....).))))))..)).))).------))).)---)....)).)))))........ ( -22.90) >DroVir_CAF1 2083 110 - 1 UACCGUUUAAUCUGUG-CGCAGGCCAUCCACCAUAUCGCUUAUCAGCUCAGCGCAACCUCAGCAAGUCGAUUGAGGAGGCGGCCA------AGCAA---UCCCAGCCUGAAUAUAAUAAC ....(((((..(((((-(...((((............(((....))).....((..(((((((.....).))))))..)))))).------.))).---...)))..)))))........ ( -27.10) >DroPse_CAF1 2260 107 - 1 -----UU----UCGUA-CACAGGUCCCACACCAUAUCGCU---CGGCCCAGCGUAACCUCAGCAAAUCGAUUGAGGCCGCCGCCGCUGCACAGCAAUCCUCCCAGCCUGAAUAUAAUAAC -----..----.....-..(((((................---.(((...(((...(((((((.....).)))))).))).)))(((....)))..........)))))........... ( -24.20) >DroGri_CAF1 2117 107 - 1 U---GUUUCGACUGUU-UUCAGGCCCUCCACCAUAUCGCUUAUCGGCACAGCGCAACCUCAGCAAGUCGAUUGAGGAGGCGGCCA------AGGAA---UCCCAGCCUGAAUAUAAUAAC .---........(((.-(((((((....................(((....(((..(((((((.....).))))))..)))))).------.((..---..)).))))))).)))..... ( -29.00) >DroWil_CAF1 22803 97 - 1 C---AUU----UUGUU-UUUAGGCCACUUACCAUAUCGCU---CUGCCCAGCGUAAUCUUAGUAAAUCAAUUGAGGAUGCUGCCA------AGCAA---UCCCAGCCUGAAUAUAAU--- .---...----((((.-(((((((.(((........((((---......)))).......)))........((.(((((((....------))).)---)))))))))))).)))).--- ( -23.66) >DroAna_CAF1 24011 93 - 1 -----U-------AUGUUACAGGUACAGCUCCCUAUCGCU---CAGCC---CGCAAUCUUAACAAAUCGAUCGAGGAUGCCGCCA------AACAG---UCCCAGCCUGAAUAUAAUAAU -----(-------(((((.(((((..(((........)))---..((.---.(((.(((..(........)..))).))).))..------.....---.....)))))))))))..... ( -16.00) >consensus _____UU____UUGUA_UACAGGCCAAACACCAUAUCGCU___CGGCCCAGCGCAACCUCAGCAAAUCGAUUGAGGAGGCCGCCA______AGCAA___UCCCAGCCUGAAUAUAAUAAC ...................(((((.............((......)).....((..((((((........))))))..))........................)))))........... (-15.48 = -14.62 + -0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:37 2006