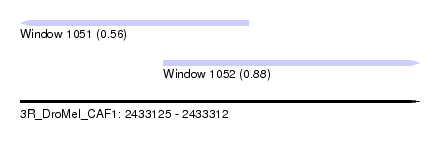
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,433,125 – 2,433,312 |
| Length | 187 |
| Max. P | 0.880931 |

| Location | 2,433,125 – 2,433,232 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.79 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -18.67 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.561031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
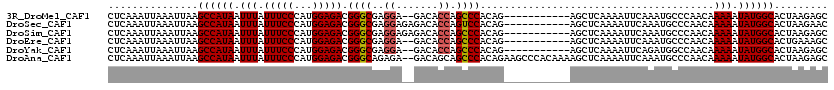
>3R_DroMel_CAF1 2433125 107 - 27905053 CUCAAAUUAAAUUAAGCCAUAAUUUAUUUCCCAUGGAGACGGGCGAGGA--GACACCAGCCCACAG-----------AGCUCAAAAUUCAAAUGCCCAACAAAAAUAUGGCACUAAGAGC ...............((((((.(((.(((((...))))).(((((.(..--..)...(((......-----------.)))...........)))))....))).))))))......... ( -22.90) >DroSec_CAF1 145748 109 - 1 CUCAAAUUAAAUUAAGCCAUAAUUUAUUUCCCAUGGAGACGGGCGAGGAGAGACACCAGUCCACAG-----------AGCUCAAAAUUCAAAUGCCCAACAAAAAUAUGGCACUAAGAAC ...............((((((.(((.(((((...))))).(((((.(((..(....)..)))...(-----------((.......)))...)))))....))).))))))......... ( -22.00) >DroSim_CAF1 140095 109 - 1 CUCAAAUUAAAUUAAGCCAUAAUUUAUUUCCCAUGGAGACGGGCGAGGAGAGACACCAGCCCACAG-----------AGCUCAAAAUUCAAAUGCCCAACAAAAAUAUGGCACUAAGAGC ...............((((((.(((.(((((...))))).(((((.((.......))(((......-----------.)))...........)))))....))).))))))......... ( -21.60) >DroEre_CAF1 141659 107 - 1 CUCAAAUUAAAUUAAGCCAUAAUUUAUUUCCCAUGGAGACGGGCGAGGA--GACACCAGCCCACAG-----------AGCUCAAAAUUCAAAUGCCCAACAAAAAUAUGGCACUGAAAGC ...............((((((.(((.(((((...))))).(((((.(..--..)...(((......-----------.)))...........)))))....))).))))))......... ( -22.90) >DroYak_CAF1 135906 107 - 1 CUCAAAUUAAAUUAAGCCAUAAUUUAUUUCCCAUGGAGACGGGCGAGGA--GACACCAGCCCACAG-----------AGCUCAAAAUUCAGAUGGCCAACAAAAAUAUGGCACUAAGAGC ...............((((((.(((.....(((((....)((((..(..--..)....))))....-----------..............))))......))).))))))......... ( -23.30) >DroAna_CAF1 141443 118 - 1 CUCAAAUUAAAUUAAGCCAUAAUUUAUUUCCCAUGGAGACGGGCAGAGA--GACAGCAGCCCACAGAAGCCCACAAAAGCUCAAAAUUCAAAUGCCCAACAAAAAUAUGGCACUAAGAGC ...............((((((.(((.(((((...))))).(((((..((--(.....(((..................))).....)))...)))))....))).))))))......... ( -21.97) >consensus CUCAAAUUAAAUUAAGCCAUAAUUUAUUUCCCAUGGAGACGGGCGAGGA__GACACCAGCCCACAG___________AGCUCAAAAUUCAAAUGCCCAACAAAAAUAUGGCACUAAGAGC ...............((((((.(((.(((((...))))).((((..((.......)).)))).......................................))).))))))......... (-18.67 = -18.87 + 0.19)
| Location | 2,433,192 – 2,433,312 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -35.79 |
| Energy contribution | -35.77 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.880931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
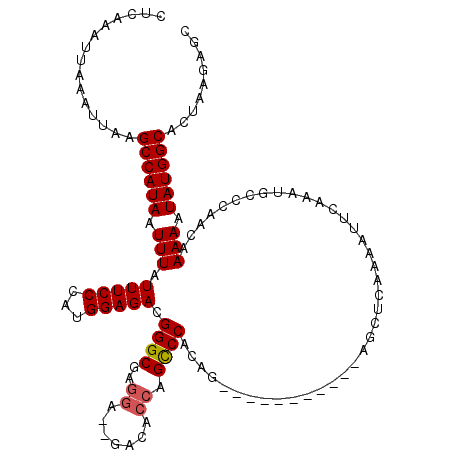
>3R_DroMel_CAF1 2433192 120 + 27905053 GUCUCCAUGGGAAAUAAAUUAUGGCUUAAUUUAAUUUGAGGUGCCUGCCCGGUCACCGCCUGUAACUGUGGCAAAAUCUGUCUGUGGUUGCUGGAUUUAUAUGCAUGCGAGUGCGGAGGC ((((((.......(((((((..(((((((......))))((((((.....)).))))))).(((((..(((((.....)))).)..)))))..)))))))..((((....)))))))))) ( -37.90) >DroSec_CAF1 145817 120 + 1 GUCUCCAUGGGAAAUAAAUUAUGGCUUAAUUUAAUUUGAGGUGCCUGCCCGGUCACCGCCUGCAACUGCGGCGAAAUCUGUCUGUGGUUGCUGGAUUUAUAUGCAUGUGAGUGCGGAGGC ((((((.......(((((((..(((((((......))))((((((.....)).))))))).(((((..(((((.....)).)))..)))))..)))))))..((((....)))))))))) ( -42.00) >DroSim_CAF1 140164 120 + 1 GUCUCCAUGGGAAAUAAAUUAUGGCUUAAUUUAAUUUGAGGUGCCUGCCCGGUCACCGCCUGCAACUGUGGCGAAAUCUGUCUGUGGUUGCUGGAUUUAUAUGCAUGUGAGUGCGGAGGC ((((((.......(((((((..(((((((......))))((((((.....)).))))))).(((((..(((((.....)))).)..)))))..)))))))..((((....)))))))))) ( -39.20) >DroEre_CAF1 141726 120 + 1 GUCUCCAUGGGAAAUAAAUUAUGGCUUAAUUUAAUUUGAGGUGCCUGCCCGGUCACCGCCUGCAACUGUGGCGAAAUCUGUCUGUGGUUGCUGGAUUUAUAUGCAUGUGAGUGCGGAGGC ((((((.......(((((((..(((((((......))))((((((.....)).))))))).(((((..(((((.....)))).)..)))))..)))))))..((((....)))))))))) ( -39.20) >DroYak_CAF1 135973 120 + 1 GUCUCCAUGGGAAAUAAAUUAUGGCUUAAUUUAAUUUGAGGUGCCUGCCCGGUCACCGCCUGCAACUGUGGUGAAAUCUGUCUGUGGUUGCUGGAUUUAUAUGCAUGUGAGUGCGGAGGC ((((((.......(((((((..(((((((......))))((((((.....)).))))))).(((((((..(.(....)...)..)))))))..)))))))..((((....)))))))))) ( -38.30) >DroAna_CAF1 141521 119 + 1 GUCUCCAUGGGAAAUAAAUUAUGGCUUAAUUUAAUUUGAGGUGCCGGCCCGGUCACCGCCUGCAA-UGUGGCGAAAUCUGUCUGUGGUUGCUGGAUUUAUAUGCCGGCGCGUGUGGAAGC (..(((((..(((.(((((((.....))))))).)))...(((((((((((((.((((((.....-...)))((......))...))).)))))........))))))))..)))))..) ( -40.00) >consensus GUCUCCAUGGGAAAUAAAUUAUGGCUUAAUUUAAUUUGAGGUGCCUGCCCGGUCACCGCCUGCAACUGUGGCGAAAUCUGUCUGUGGUUGCUGGAUUUAUAUGCAUGUGAGUGCGGAGGC ((((((.......(((((((..(((((((......))))((((((.....)).))))))).(((((..(((((.....)).)))..)))))..)))))))..((((....)))))))))) (-35.79 = -35.77 + -0.03)

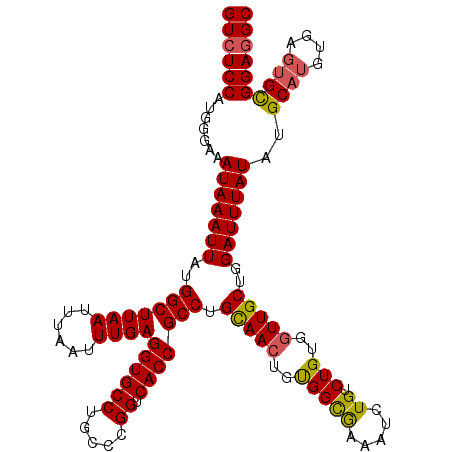
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:44 2006