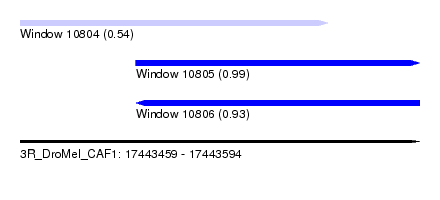
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,443,459 – 17,443,594 |
| Length | 135 |
| Max. P | 0.992710 |

| Location | 17,443,459 – 17,443,563 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.03 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -11.80 |
| Energy contribution | -11.68 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17443459 104 + 27905053 -ACUCAGCAUUUUGGUCACUCUAAUUACGAGCUCCAAAAUAAAAUAAAAAACAGCACUGUCAUGACUUUUUGACAACCGCUGAAAGUGUGGUGUCCACCAU---------------ACAU -..((((((((((((...(((.......)))..))))))).................(((((........)))))...)))))..(((((((....)))))---------------)).. ( -30.30) >DroSec_CAF1 44950 104 + 1 -GCUCAGCAUUUUGGUCACUCUAAUUACGAGCUCCAAAAUAAUAUAAAAAACAGCAGUGUCAUGCCUUUUUGACAACCACUGAAAAUGUGGUGUCCAACAU---------------ACAU -.......(((((((...(((.......)))..))))))).......((((..(((......)))..))))((((.((((.......))))))))......---------------.... ( -20.50) >DroSim_CAF1 42152 104 + 1 -GCUCAGCAUUUUGGUCACUCUAAUUACGAGCUCCAAAAUAAUAUAAAAAACAGCAGUGUCAUGCCAUUUUGACAACCCCUGAAAAUGUGGUGUCCAACAU---------------ACAU -..((((.(((((((...(((.......)))..))))))).................(((((........)))))....))))..(((((((.....)).)---------------)))) ( -18.50) >DroEre_CAF1 44374 119 + 1 AGCUCAGCUAUUUGGUCACACUAAUUACGAGCUCCAAAAUAAAAUC-AAAACUGCACUGUCAUGACUUUUUGACAAAUGCUGAAAAUGUGGUGUUCAACUUAGUUGUUAAUUAAACACGU ....((((((.((((.....))))....((((.(((........((-(.....(((.(((((........)))))..)))))).....))).))))....)))))).............. ( -20.22) >DroYak_CAF1 44834 119 + 1 UGCACAGCUAA-UGGUCACUCUAACUACGAGCUCCAAAAUAAAAUUGAAAACUGCAAUGUCAUGACUUUUUGACAAACACUAAAAUUAUGGUGUCCAACUUAGUUGUUAAUUAAACACGU ...((((((((-(((.((((.(((......((.....................))..(((((........)))))..........))).)))).)))..))))))))............. ( -21.40) >consensus _GCUCAGCAUUUUGGUCACUCUAAUUACGAGCUCCAAAAUAAAAUAAAAAACAGCACUGUCAUGACUUUUUGACAACCACUGAAAAUGUGGUGUCCAACAU_______________ACAU ............(((.((((.((.......((.....................))..(((((........)))))...........)).)))).)))....................... (-11.80 = -11.68 + -0.12)


| Location | 17,443,498 – 17,443,594 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.98 |
| Mean single sequence MFE | -18.99 |
| Consensus MFE | -9.94 |
| Energy contribution | -10.66 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17443498 96 + 27905053 AAAAUAAAAAACAGCACUGUCAUGACUUUUUGACAACCGCUGAAAGUGUGGUGUCCACCAU---------------ACAUUAAA-AA--------ACAACACAUUUUCCAAAGUAUAACU ...........((((..(((((........)))))...))))...(((((((....)))))---------------))......-..--------......................... ( -20.20) >DroSec_CAF1 44989 97 + 1 AAUAUAAAAAACAGCAGUGUCAUGCCUUUUUGACAACCACUGAAAAUGUGGUGUCCAACAU---------------ACAUUAAAAAA--------ACAACACAUUUUCCAAAGUAUAACU .................(((((........)))))...((((((((((((.(((.......---------------...........--------))).)))))))))...)))...... ( -14.77) >DroSim_CAF1 42191 97 + 1 AAUAUAAAAAACAGCAGUGUCAUGCCAUUUUGACAACCCCUGAAAAUGUGGUGUCCAACAU---------------ACAUUAAAAAA--------ACAACACAUUUUCCAAAGUAUAACU .............((..(((((........)))))......(((((((((.(((.......---------------...........--------))).)))))))))....))...... ( -14.77) >DroEre_CAF1 44414 118 + 1 AAAAUC-AAAACUGCACUGUCAUGACUUUUUGACAAAUGCUGAAAAUGUGGUGUUCAACUUAGUUGUUAAUUAAACACGUUAAAAAAGGGCGUAGACCACACAUUUUCCAA-AAAUGAUU ..((((-(.....(((.(((((........)))))..))).(((((((((.((.((......(((........)))(((((.......))))).)).)))))))))))...-...))))) ( -25.60) >DroYak_CAF1 44873 119 + 1 AAAAUUGAAAACUGCAAUGUCAUGACUUUUUGACAAACACUAAAAUUAUGGUGUCCAACUUAGUUGUUAAUUAAACACGUUAAAAAAGGGCGUAAACAAC-CGUUUUCCAACAAAUGAUU ..((((((.(((((...(((((........))))).((((((......))))))......))))).))))))......(((..((((.((.........)-).))))..)))........ ( -19.60) >consensus AAAAUAAAAAACAGCACUGUCAUGACUUUUUGACAACCACUGAAAAUGUGGUGUCCAACAU_______________ACAUUAAAAAA________ACAACACAUUUUCCAAAGUAUAACU .................(((((........)))))......(((((((((.(((.........................................))).)))))))))............ ( -9.94 = -10.66 + 0.72)

| Location | 17,443,498 – 17,443,594 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.98 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -12.24 |
| Energy contribution | -13.04 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17443498 96 - 27905053 AGUUAUACUUUGGAAAAUGUGUUGU--------UU-UUUAAUGU---------------AUGGUGGACACCACACUUUCAGCGGUUGUCAAAAAGUCAUGACAGUGCUGUUUUUUAUUUU ...(((((.(((((((((.....))--------))-))))).))---------------)))((((...)))).....(((((.(.((((........))))).)))))........... ( -21.30) >DroSec_CAF1 44989 97 - 1 AGUUAUACUUUGGAAAAUGUGUUGU--------UUUUUUAAUGU---------------AUGUUGGACACCACAUUUUCAGUGGUUGUCAAAAAGGCAUGACACUGCUGUUUUUUAUAUU ............(((((((((.(((--------((..(......---------------..)..))))).)))))))))((..((.((((........))))))..))............ ( -22.60) >DroSim_CAF1 42191 97 - 1 AGUUAUACUUUGGAAAAUGUGUUGU--------UUUUUUAAUGU---------------AUGUUGGACACCACAUUUUCAGGGGUUGUCAAAAUGGCAUGACACUGCUGUUUUUUAUAUU .(....((((..(((((((((.(((--------((..(......---------------..)..))))).)))))))))..))))...)(((((((((......)))))))))....... ( -22.90) >DroEre_CAF1 44414 118 - 1 AAUCAUUU-UUGGAAAAUGUGUGGUCUACGCCCUUUUUUAACGUGUUUAAUUAACAACUAAGUUGAACACCACAUUUUCAGCAUUUGUCAAAAAGUCAUGACAGUGCAGUUUU-GAUUUU (((((...-...(((((((((.(((....)))..........((((((((((........))))))))))))))))))).(((((.((((........))))))))).....)-)))).. ( -31.20) >DroYak_CAF1 44873 119 - 1 AAUCAUUUGUUGGAAAACG-GUUGUUUACGCCCUUUUUUAACGUGUUUAAUUAACAACUAAGUUGGACACCAUAAUUUUAGUGUUUGUCAAAAAGUCAUGACAUUGCAGUUUUCAAUUUU ........(((((((((.(-((.......))).)))))))))((((((((((........))))))))))...........(((.(((((........)))))..)))............ ( -23.30) >consensus AGUUAUACUUUGGAAAAUGUGUUGU________UUUUUUAAUGU_______________AUGUUGGACACCACAUUUUCAGCGGUUGUCAAAAAGUCAUGACACUGCUGUUUUUUAUUUU ............(((((((((.(((.........................................))).))))))))).(((((.((((........)))))))))............. (-12.24 = -13.04 + 0.80)

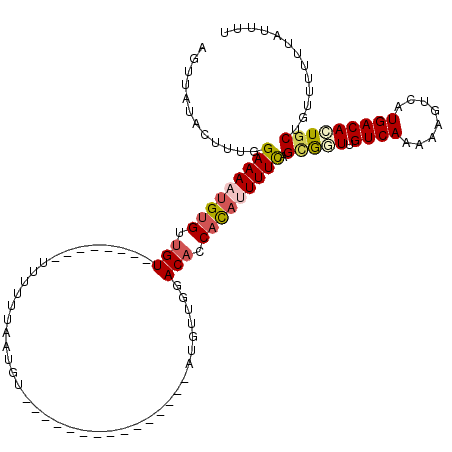
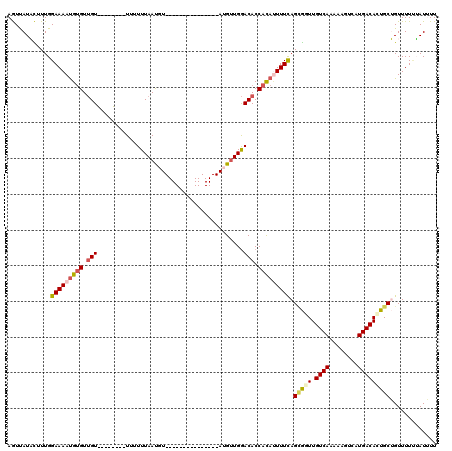
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:31 2006