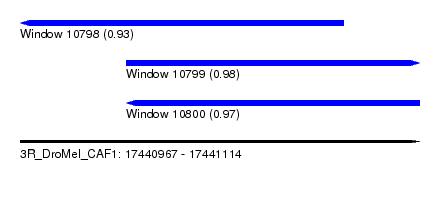
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,440,967 – 17,441,114 |
| Length | 147 |
| Max. P | 0.981924 |

| Location | 17,440,967 – 17,441,086 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -31.76 |
| Energy contribution | -31.80 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17440967 119 - 27905053 ACCCAGCCGCAACAGCCAUGGCAAUAACAAUAGUGCUGGCGACAGCGGGAACAAAAGUAACGCAUGGAGCCAGCCCCUCCACGCGCACACUUGGAGCCAACAACACAUGACAUGGUCGA ............(.((((((.((...........((((((..(((((...((....))..))).))..))))))..(((((.(......).)))))...........)).)))))).). ( -33.70) >DroSec_CAF1 42438 119 - 1 AACCAGCCGCAACAGCCAUGGCAAUAACACUAGUGCUGGCGACAGCGGGAACAAAAGUAACGCAUGGAGCCAGCCCCUCCACGCGCACACUUGGAGCCAACAACACAUGACAUGGUCGA ............(.((((((.((...........((((((..(((((...((....))..))).))..))))))..(((((.(......).)))))...........)).)))))).). ( -33.70) >DroSim_CAF1 39560 118 - 1 AACCAGCCGCAACAGCCAUGGCAAUAACAAUAGUGCUGGCGACAGCGGGAACAA-AGUAACGCAUGGAGCCAGCCCCUCCACGCGCACACUUGGAGCCAACAACACAUGACAUGGUCGA ............(.((((((.((...........((((((..(((((..(....-..)..))).))..))))))..(((((.(......).)))))...........)).)))))).). ( -33.30) >DroEre_CAF1 41916 115 - 1 ACCCAGGCGCAACAGCCAUGGCAAUAACAAUAGUGCUGGCGACAGCAGGAACAAAAGUAACGCAUGGAG----CCCCUCCACGCGCGCACUUGGAGCCAACAACACAUGACAUGGUCGG ..((((((......))).((((...........(((((....))))).......((((..(((.(((((----...))))).)))...))))...)))).............))).... ( -34.50) >DroYak_CAF1 42365 115 - 1 ACCCAGCCGCAACAGCCAUGGCAAUAACAAUAGUGCUGGCGACAGCGGGAACAAAAGUAACGCAUGGAG----CCCCUCCACGCGCACACUUGGAACCAACAACACAUGACAUGGUCGA ............(.((((((.((...........((((....))))((...(((..((..(((.(((((----...))))).))).))..)))...)).........)).)))))).). ( -31.50) >consensus ACCCAGCCGCAACAGCCAUGGCAAUAACAAUAGUGCUGGCGACAGCGGGAACAAAAGUAACGCAUGGAGCCAGCCCCUCCACGCGCACACUUGGAGCCAACAACACAUGACAUGGUCGA ............(.((((((.((..........(((((....)))))((.....((((..(((.(((((.......))))).)))...))))....)).........)).)))))).). (-31.76 = -31.80 + 0.04)

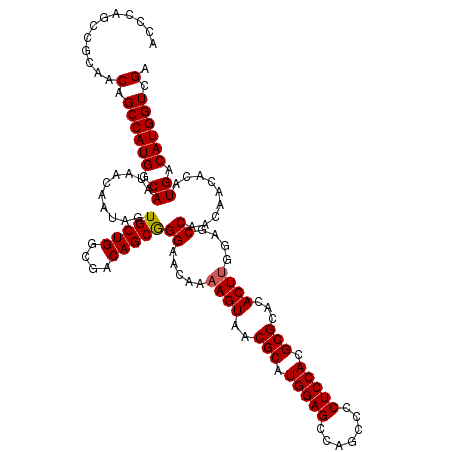
| Location | 17,441,006 – 17,441,114 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 95.35 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -30.54 |
| Energy contribution | -32.42 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17441006 108 + 27905053 GGAGGGGCUGGCUCCAUGCGUUACUUUUGUUCCCGCUGUCGCCAGCACUAUUGUUAUUGCCAUGGCUGUUGCGGCUGGGUAUUAUUUGCUUGGACCUUAGACCCUCUU ((((((((((((..((.(((..((....))...)))))..)))))).(((..((.((.((....)).)).))(((..((((.....))))..).)).))).)))))). ( -41.50) >DroSec_CAF1 42477 108 + 1 GGAGGGGCUGGCUCCAUGCGUUACUUUUGUUCCCGCUGUCGCCAGCACUAGUGUUAUUGCCAUGGCUGUUGCGGCUGGUUAUUAUUUACUUGGACCUUAGACCCUCUU ((((((((((((..((.(((..((....))...)))))..))))))((((((((.((.((....)).)).)).))))))......................)))))). ( -37.50) >DroSim_CAF1 39599 107 + 1 GGAGGGGCUGGCUCCAUGCGUUACU-UUGUUCCCGCUGUCGCCAGCACUAUUGUUAUUGCCAUGGCUGUUGCGGCUGGUUAUUAUUUACUUGGACCUUAGACCCUCUU ((((((((((((..((.(((.....-.......)))))..)))))).(((..(((...(((((.((....)).).)))).............)))..))).)))))). ( -34.49) >DroEre_CAF1 41955 104 + 1 GGAGGGG----CUCCAUGCGUUACUUUUGUUCCUGCUGUCGCCAGCACUAUUGUUAUUGCCAUGGCUGUUGCGCCUGGGUAUUAUUUACUUGGACCUUAGACCCUCUU ((((((.----......((((.((.........(((((....)))))...........((....)).)).))))(..((((.....))))..)........)))))). ( -29.30) >DroYak_CAF1 42404 104 + 1 GGAGGGG----CUCCAUGCGUUACUUUUGUUCCCGCUGUCGCCAGCACUAUUGUUAUUGCCAUGGCUGUUGCGGCUGGGUAUUAUUUACUUGGACCUUAGACCCUCUU (((((((----(..((.(((..((....))...)))....(((((((.((....)).)))..))))))..))(((..((((.....))))..).)).....)))))). ( -32.60) >consensus GGAGGGGCUGGCUCCAUGCGUUACUUUUGUUCCCGCUGUCGCCAGCACUAUUGUUAUUGCCAUGGCUGUUGCGGCUGGGUAUUAUUUACUUGGACCUUAGACCCUCUU ((((((((((((..((.(((..((....))...)))))..)))))).(((..((.((.((....)).)).))(((..((((.....))))..).)).))).)))))). (-30.54 = -32.42 + 1.88)

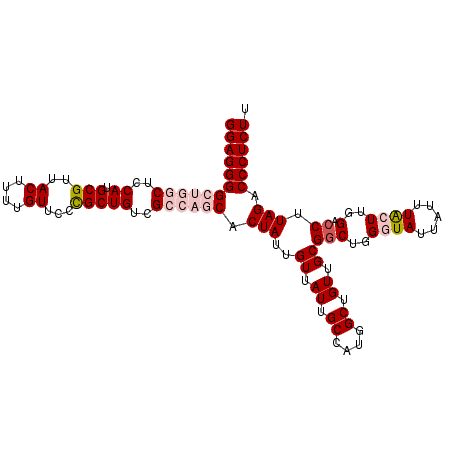
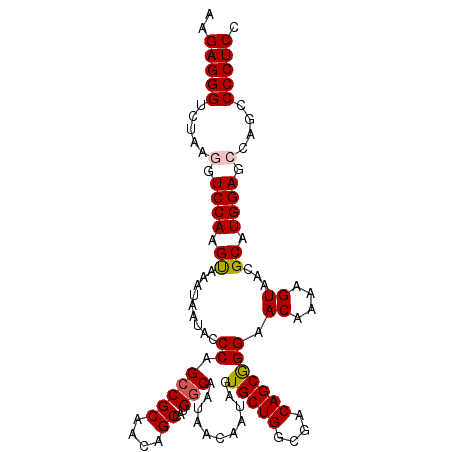
| Location | 17,441,006 – 17,441,114 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 95.35 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -25.88 |
| Energy contribution | -26.16 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17441006 108 - 27905053 AAGAGGGUCUAAGGUCCAAGCAAAUAAUACCCAGCCGCAACAGCCAUGGCAAUAACAAUAGUGCUGGCGACAGCGGGAACAAAAGUAACGCAUGGAGCCAGCCCCUCC ..(((((.((..((((((.((........(((.(((((....))...)))............((((....))))))).((....))...)).)))).)))).))))). ( -36.20) >DroSec_CAF1 42477 108 - 1 AAGAGGGUCUAAGGUCCAAGUAAAUAAUAACCAGCCGCAACAGCCAUGGCAAUAACACUAGUGCUGGCGACAGCGGGAACAAAAGUAACGCAUGGAGCCAGCCCCUCC ..(((((.(((.(((..............))).(((((....))...)))........))).((((((..(((((...((....))..))).))..))))))))))). ( -32.94) >DroSim_CAF1 39599 107 - 1 AAGAGGGUCUAAGGUCCAAGUAAAUAAUAACCAGCCGCAACAGCCAUGGCAAUAACAAUAGUGCUGGCGACAGCGGGAACAA-AGUAACGCAUGGAGCCAGCCCCUCC ..(((((.(((.(((..............))).(((((....))...)))........))).((((((..(((((..(....-..)..))).))..))))))))))). ( -32.74) >DroEre_CAF1 41955 104 - 1 AAGAGGGUCUAAGGUCCAAGUAAAUAAUACCCAGGCGCAACAGCCAUGGCAAUAACAAUAGUGCUGGCGACAGCAGGAACAAAAGUAACGCAUGGAG----CCCCUCC ..(((((.....(.((((.((.........((((((......))).)))............(((((....)))))..............)).)))).----)))))). ( -30.60) >DroYak_CAF1 42404 104 - 1 AAGAGGGUCUAAGGUCCAAGUAAAUAAUACCCAGCCGCAACAGCCAUGGCAAUAACAAUAGUGCUGGCGACAGCGGGAACAAAAGUAACGCAUGGAG----CCCCUCC ..(((((.....(.((((.((........(((.(((((....))...)))............((((....))))))).((....))...)).)))).----)))))). ( -30.70) >consensus AAGAGGGUCUAAGGUCCAAGUAAAUAAUACCCAGCCGCAACAGCCAUGGCAAUAACAAUAGUGCUGGCGACAGCGGGAACAAAAGUAACGCAUGGAGCCAGCCCCUCC ..(((((.....(.((((.((.........((.(((((....))...)))...........(((((....))))))).((....))...)).)))).)....))))). (-25.88 = -26.16 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:26 2006