
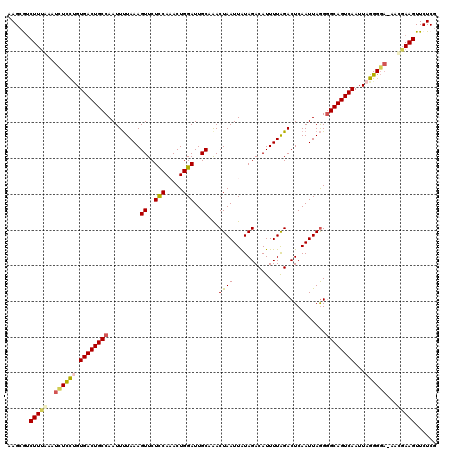
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,433,832 – 17,433,951 |
| Length | 119 |
| Max. P | 0.981533 |

| Location | 17,433,832 – 17,433,951 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17433832 119 + 27905053 AAGCGUCUUCGAAUUUCCUGUGACUGCCAAUUUUAAAGUUCUCCAAACUGGAUUGCAAACUAAUUAUAGACAUUUUGGACUCAAUUAGGGGCAGUCAAUUAGGGGA-AACGAAGUUCUCG ......(((((..(..((((((((((((.........((..(((.....)))..))...((((((.....(.....).....)))))).))))))))..))))..)-..)))))...... ( -34.10) >DroSec_CAF1 37824 119 + 1 AAGCGUCUUUAAAUCUCCCGUGACUGCCAAUUUUAAAGUUCUCCACACUGGAUUGCAAACUAAUUAUAGACAUUUUAGACUCAAUUAGGGGCAGUCAAUUAGGGGA-AACUAAGUUCUCG ......(((....(((((..((((((((.........((..(((.....)))..))...((((((..((.(......).)).)))))).))))))))....)))))-....)))...... ( -28.60) >DroSim_CAF1 34965 120 + 1 AAGCGUCUUUAAAUCUCCUGUGACUGCCAAUUUUAAAGUUCUCCACACUGGAUUGCAAACUAAUUAUAGACAUUUUAGACUCAAUUAGGGGCAGUCAAUUAGGGGAAAACGAAGUUCUCG ......((((...(((((((((((((((.........((..(((.....)))..))...((((((..((.(......).)).)))))).))))))))..)))))))....))))...... ( -30.70) >DroEre_CAF1 37570 119 + 1 GAGGACCUUUUAAUCUCCAGUGACUGCCAAUUUUAAAGUUCUCCAAACUGGAUUGCAGACUAAUUAUAGACAUUUUAGACUCAAUUAGGGGCAGUCAAUUAGGGGA-AAAGAAGUUCUCG ((((((.((((..(((((..((((((((.........((..(((.....)))..))...((((((..((.(......).)).)))))).))))))))....)))))-..)))))))))). ( -37.40) >DroYak_CAF1 37986 116 + 1 GAGCAUCUUUUUAUCUUUUGUGACUGCCAAUUCUAAAGUUCUUCAAACUGGAUUGCAAACUAAUUGUAGACAUUUUAGACUCAAUUAAGUGCAGUCAAUUAGGGU----AGAAGUUCUCG (((....((((((((((...(((((((((((((...((((.....))))))))))...((((((((.((.(......).))))))).))))))))))...)))))----)))))..))). ( -28.70) >consensus AAGCGUCUUUAAAUCUCCUGUGACUGCCAAUUUUAAAGUUCUCCAAACUGGAUUGCAAACUAAUUAUAGACAUUUUAGACUCAAUUAGGGGCAGUCAAUUAGGGGA_AACGAAGUUCUCG ......(((((..((((((.((((((((.........((..(((.....)))..))...((((...........))))...........))))))))...))))))...)))))...... (-25.08 = -25.64 + 0.56)


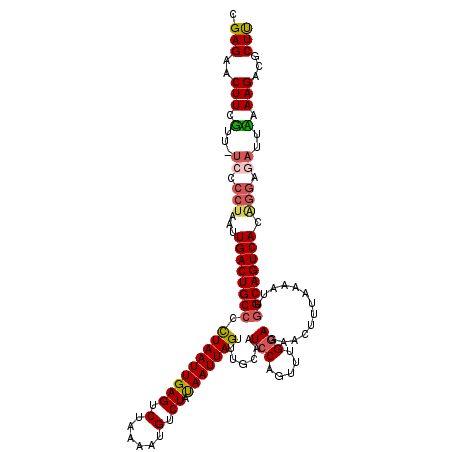
| Location | 17,433,832 – 17,433,951 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.28 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17433832 119 - 27905053 CGAGAACUUCGUU-UCCCCUAAUUGACUGCCCCUAAUUGAGUCCAAAAUGUCUAUAAUUAGUUUGCAAUCCAGUUUGGAGAACUUUAAAAUUGGCAGUCACAGGAAAUUCGAAGACGCUU .(((..(((((..-(..(((...((((((((.(((((((((.(......).)).))))))).......(((.....))).............)))))))).)))..)..)))))...))) ( -29.90) >DroSec_CAF1 37824 119 - 1 CGAGAACUUAGUU-UCCCCUAAUUGACUGCCCCUAAUUGAGUCUAAAAUGUCUAUAAUUAGUUUGCAAUCCAGUGUGGAGAACUUUAAAAUUGGCAGUCACGGGAGAUUUAAAGACGCUU .(((..(((((((-((((.....((((((((.(((((((((.(......).)).)))))))((..((......))..)).............)))))))).))))))))..)))...))) ( -31.00) >DroSim_CAF1 34965 120 - 1 CGAGAACUUCGUUUUCCCCUAAUUGACUGCCCCUAAUUGAGUCUAAAAUGUCUAUAAUUAGUUUGCAAUCCAGUGUGGAGAACUUUAAAAUUGGCAGUCACAGGAGAUUUAAAGACGCUU .((((((...)))))).(((...((((((((.(((((((((.(......).)).)))))))((..((......))..)).............)))))))).)))................ ( -27.10) >DroEre_CAF1 37570 119 - 1 CGAGAACUUCUUU-UCCCCUAAUUGACUGCCCCUAAUUGAGUCUAAAAUGUCUAUAAUUAGUCUGCAAUCCAGUUUGGAGAACUUUAAAAUUGGCAGUCACUGGAGAUUAAAAGGUCCUC .(((.((((.((.-((.((....((((((((.(((((((((.(......).)).)))))))(((.(((......))).)))...........))))))))..)).))..)).)))).))) ( -31.10) >DroYak_CAF1 37986 116 - 1 CGAGAACUUCU----ACCCUAAUUGACUGCACUUAAUUGAGUCUAAAAUGUCUACAAUUAGUUUGCAAUCCAGUUUGAAGAACUUUAGAAUUGGCAGUCACAAAAGAUAAAAAGAUGCUC .(((..(((..----...((...(((((((..(((((((((.(......).)).))))))).........(((((((((....))))).)))))))))))....)).....)))...))) ( -20.50) >consensus CGAGAACUUCGUU_UCCCCUAAUUGACUGCCCCUAAUUGAGUCUAAAAUGUCUAUAAUUAGUUUGCAAUCCAGUUUGGAGAACUUUAAAAUUGGCAGUCACAGGAGAUUAAAAGACGCUU .(((..(((.(...((.(((...((((((((.(((((((((.(......).)).))))))).......(((.....))).............)))))))).))).))..).)))...))) (-20.92 = -21.28 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:22 2006