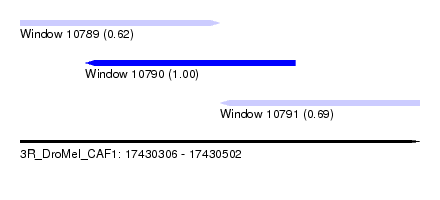
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,430,306 – 17,430,502 |
| Length | 196 |
| Max. P | 0.995396 |

| Location | 17,430,306 – 17,430,404 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 98.64 |
| Mean single sequence MFE | -18.84 |
| Consensus MFE | -18.73 |
| Energy contribution | -18.51 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17430306 98 + 27905053 ACUUAAUGGGCAUGAAAACCGUCUGCGCCAGAAACAACAACAAAAGCAAGAAAAACAAGCGCAGUUCAAGCAUCAAAAAGCUGCCGUCACGCCCCUCU .......((((.(((.....(.((((((..............................))))))..).(((........)))....))).)))).... ( -19.51) >DroSec_CAF1 34194 98 + 1 ACCUAAUGGGUAUGAAAACCGUCUGCGCCAGAAACAACAACAAAAGCAAGAAAAACAAGCGCAGUUCAAGCAUCAAAAAGCUGCCGUCACGCCCCUCU .......(((..(((.....(.((((((..............................))))))..).(((........)))....)))..))).... ( -18.51) >DroSim_CAF1 31334 98 + 1 ACUUAAUGGGUAUGAAAACCGUCUGCGCCAGAAACAACAACAAAAGCAAGAAAAACAAGCGCAGUUCAAGCAUCAAAAAGCUGCCGUCACGCCCCUCU .......(((..(((.....(.((((((..............................))))))..).(((........)))....)))..))).... ( -18.51) >consensus ACUUAAUGGGUAUGAAAACCGUCUGCGCCAGAAACAACAACAAAAGCAAGAAAAACAAGCGCAGUUCAAGCAUCAAAAAGCUGCCGUCACGCCCCUCU .......((((.(((.....(.((((((..............................))))))..).(((........)))....))).)))).... (-18.73 = -18.51 + -0.22)

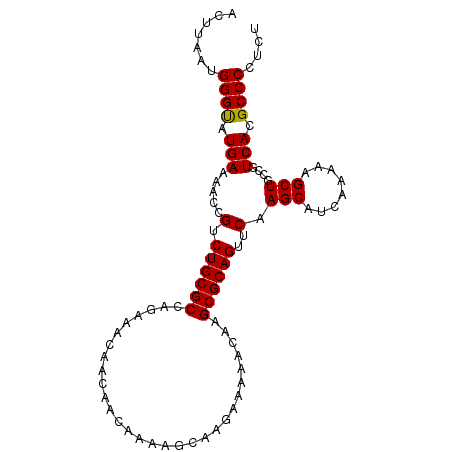
| Location | 17,430,338 – 17,430,441 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -39.64 |
| Consensus MFE | -32.66 |
| Energy contribution | -33.10 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17430338 103 - 27905053 GCAGGGAAGGCAGGGGGUGGUGAGCGGCAAG--CACUCAAGAGGGGCGUGACGGCAGCUUUUUGAUGCUUGAACUGCGCUUGUUUUUCUUGCUUUUGUUGUUGUU ((((.(((((((((..(.((..((((.((((--((.((((((((.((......))..)))))))))))))).....))))..)))..))))))))).)))).... ( -43.20) >DroSec_CAF1 34226 104 - 1 GCAGGGAAGGCAGG-GGUGGUGAGCGGCAAGCGCACUCAAGAGGGGCGUGACGGCAGCUUUUUGAUGCUUGAACUGCGCUUGUUUUUCUUGCUUUUGUUGUUGUU ((((.(((((((((-((..(..((((.((((((...((((((((.((......))..)))))))))))))).....))))..)..))))))))))).)))).... ( -41.60) >DroSim_CAF1 31366 104 - 1 GCAGGGAGGGCAGG-GGUGGUGAGCGGCAAGCGCACUCAAGAGGGGCGUGACGGCAGCUUUUUGAUGCUUGAACUGCGCUUGUUUUUCUUGCUUUUGUUGUUGUU ((((.(((((((((-((..(..((((.((((((...((((((((.((......))..)))))))))))))).....))))..)..))))))))))).)))).... ( -41.20) >DroEre_CAF1 33969 104 - 1 GCAGAGGAGGCAGG-GGUGGUGAGCGGCAAGCUCACUCAAGAGGGGCGUGACGGCAGCUUUUUGAUGCUUGAACUGCGCUUGUUUUUCUUGCUUUUGUUGUUGUU ((((..((((((((-((..(..((((.(((((....((((((((.((......))..)))))))).))))).....))))..)..))))))))))..)))).... ( -39.30) >DroYak_CAF1 34602 104 - 1 GCAGAGGAGGCAGG-GGUAGUGCGCGGCAAGCGCACUCAAGAGGGGCGUGAUGGCAGCUUUUUGAUGCUUGAACUUCGCUUGUUUUUCUUGGUUUUGUUGUUAUU ((((..(((.((((-((.(((((((.....))))))))((((.(((((....((((.(.....).)))).......))))).))))))))).)))..)))).... ( -32.90) >consensus GCAGGGAAGGCAGG_GGUGGUGAGCGGCAAGCGCACUCAAGAGGGGCGUGACGGCAGCUUUUUGAUGCUUGAACUGCGCUUGUUUUUCUUGCUUUUGUUGUUGUU ((((.(((((((((....((..((((.((((((...((((((((.((......))..)))))))))))))).....))))..))...))))))))).)))).... (-32.66 = -33.10 + 0.44)


| Location | 17,430,404 – 17,430,502 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.02 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -29.02 |
| Energy contribution | -28.66 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17430404 98 - 27905053 GCGGGCUGCGGUGGAGCGGUAAAGCG--------AGAGAGACGCUGUAUGUCUGGCCCCGUUUGAUGGGGCAGGGAAGGCAGGGGGUGGUGAGCGGCAAG--CACUCA ((..(((((......)))))...))(--------((.....((((.(.(((((.(((((((...))))))).....))))).).))))....((.....)--).))). ( -38.00) >DroSec_CAF1 34292 107 - 1 GCGGGCUGCGGUGGAGCGGUAGAGCGAGAGAGCGAGAGAGACGCUGUAUGUCUGGCCUCGUUUGAUGGGGCAGGGAAGGCAGG-GGUGGUGAGCGGCAAGCGCACUCA ((..(((((......)))))...))..(((.(((.......(.((((...(((.(((((((...)))))))..)))..)))).-)((.(....).))...))).))). ( -39.30) >DroSim_CAF1 31432 99 - 1 GCGGGCUGCGUUGGAGCGGUAGAGCG--------AGAGAGACGCUGUAUGUCUGGCCUCGUUUGAUGGGGCAGGGAGGGCAGG-GGUGGUGAGCGGCAAGCGCACUCA ((..(((((......)))))...))(--------((.....(((..(.(((((.(((((((...))))))).....)))))..-.)..))).(((.....))).))). ( -36.80) >DroEre_CAF1 34035 98 - 1 GUGGG-AGCGGGGGAGCGGUAGAGCG--------AGAGAGACGCUGUAGGUCUGGCCUCGUUUGAUGGGGCAGAGGAGGCAGG-GGUGGUGAGCGGCAAGCUCACUCA ...((-(.(...(.((((......(.--------...)...)))).).).))).(((((.((((......)))).)))))...-...(((((((.....))))))).. ( -34.60) >DroYak_CAF1 34668 98 - 1 GUGGG-AGCUGUGGAGCAGCAAAGCG--------AGAGAGACGCUGUAGGUCUGGCCUCGUAUGGUGGGGCAGAGGAGGCAGG-GGUAGUGCGCGGCAAGCGCACUCA .....-.((((.....))))......--------.......(.((((...(((.((((((.....))))))..)))..)))).-)(.(((((((.....)))))))). ( -35.80) >consensus GCGGGCUGCGGUGGAGCGGUAGAGCG________AGAGAGACGCUGUAUGUCUGGCCUCGUUUGAUGGGGCAGGGAAGGCAGG_GGUGGUGAGCGGCAAGCGCACUCA ((..(((((......)))))...))..................((((...(((.(((((((...)))))))..)))..)))).....((((.((.....)).)))).. (-29.02 = -28.66 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:17 2006