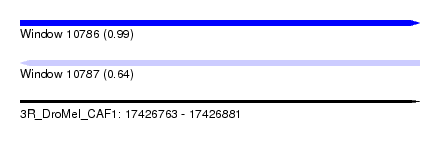
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,426,763 – 17,426,881 |
| Length | 118 |
| Max. P | 0.989284 |

| Location | 17,426,763 – 17,426,881 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -38.85 |
| Consensus MFE | -29.44 |
| Energy contribution | -32.48 |
| Covariance contribution | 3.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
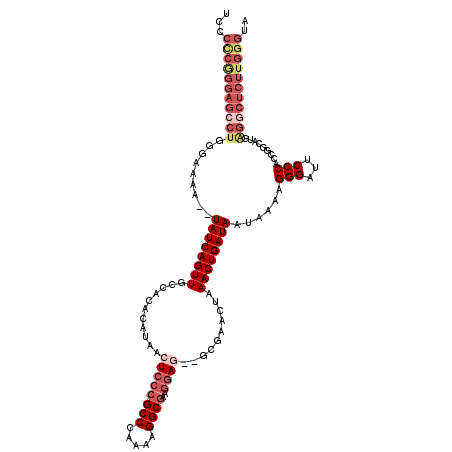
>3R_DroMel_CAF1 17426763 118 + 27905053 UCCCCCGGGAGCCUGGGAAAAAUUAUCAGUUGCCACACAUAACUCCCGCCCAAAAGGCGAAGGAG--GCGAACUAAACUGAUAAUAAAAGGGAUUCCCACCGGCAUAAGGCUCUUGGGUA ...(((((((((((.((....((((((((((...........((((((((.....))))..))))--........))))))))))....(((...))).))......))))))))))).. ( -44.51) >DroSec_CAF1 30658 118 + 1 UCCCCCGGGAGCCUGGGAAAA--UAUCAGUUGCCACACAUAACUCCCGCCCAAAAGGCGAAGGAGGGGCGAACUAAACUGAUAAUAAAAGGGAUUCCCGCCGGCAUGAGGCUCUUGGGUA ...(((((((((((.(.....--(((((((((((........((((((((.....))))..)))).)))......))))))))......(((...))).......).))))))))))).. ( -44.80) >DroSim_CAF1 28001 118 + 1 UCCCCCGGGAGCCUGGGAAAA--UAUCAGUUGCCACACAUAACUCCCGCCCAAAAGGCGAAGGAGGGGCGAACUAAACUGAUAAUAAAAGGGAUUCCCACCGGCAUGGGGCUCUUGGGUA ...((((((((((((((((..--(((((((((((........((((((((.....))))..)))).)))......))))))))..........))))))((.....)))))))))))).. ( -45.10) >DroEre_CAF1 30419 116 + 1 UCCCUCAGGAGCCUGGGAAAA--UAUCAGUUGCCACACAUAACUCCCGCCCAAAAGGCGAAGGAU--GCGAACUAAACUGAUAAUAAAAGGGAUUCCCACAGGCGUGAGACUCUUGGGUA ...((((((((((((......--((((((((....(.(((..((..((((.....))))..))))--).).....))))))))......(((...))).))))(....).)))))))).. ( -34.80) >DroYak_CAF1 28515 97 + 1 UCCCACGGGAGCCUGGGAAAA--UAUCAGUUGCCACACAGAACUCGAGCCCAAAAGGCGAAGAAU--GCGAACUAAACUGAUAAUAAAAGGGAUUCCCACA------------------- ......((((((((.......--((((((((....(.((....((..(((.....)))...)).)--).).....))))))))......))).)))))...------------------- ( -25.02) >consensus UCCCCCGGGAGCCUGGGAAAA__UAUCAGUUGCCACACAUAACUCCCGCCCAAAAGGCGAAGGAG__GCGAACUAAACUGAUAAUAAAAGGGAUUCCCACCGGCAUGAGGCUCUUGGGUA ...(((((((((((.........((((((((...........((((((((.....))))..))))..........))))))))......(((...))).........))))))))))).. (-29.44 = -32.48 + 3.04)


| Location | 17,426,763 – 17,426,881 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -37.86 |
| Consensus MFE | -27.98 |
| Energy contribution | -30.30 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
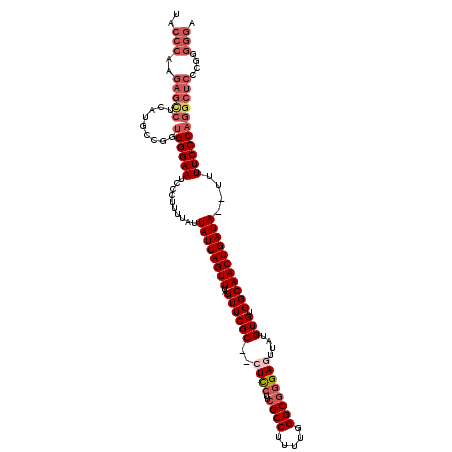
>3R_DroMel_CAF1 17426763 118 - 27905053 UACCCAAGAGCCUUAUGCCGGUGGGAAUCCCUUUUAUUAUCAGUUUAGUUCGC--CUCCUUCGCCUUUUGGGCGGGAGUUAUGUGUGGCAACUGAUAAUUUUUCCCAGGCUCCCGGGGGA ..(((..((((((.........(((((........((((((((((..((((((--((((..((((.....))))))))....))).)))))))))))))..)))))))))))...))).. ( -41.70) >DroSec_CAF1 30658 118 - 1 UACCCAAGAGCCUCAUGCCGGCGGGAAUCCCUUUUAUUAUCAGUUUAGUUCGCCCCUCCUUCGCCUUUUGGGCGGGAGUUAUGUGUGGCAACUGAUA--UUUUCCCAGGCUCCCGGGGGA ..(((..((((((...(....)(((((..........((((((((..((((((..((((..((((.....))))))))....))).)))))))))))--..)))))))))))...))).. ( -41.20) >DroSim_CAF1 28001 118 - 1 UACCCAAGAGCCCCAUGCCGGUGGGAAUCCCUUUUAUUAUCAGUUUAGUUCGCCCCUCCUUCGCCUUUUGGGCGGGAGUUAUGUGUGGCAACUGAUA--UUUUCCCAGGCUCCCGGGGGA ..(((..(((((((.....))((((((..........((((((((..((((((..((((..((((.....))))))))....))).)))))))))))--..)))))))))))...))).. ( -42.40) >DroEre_CAF1 30419 116 - 1 UACCCAAGAGUCUCACGCCUGUGGGAAUCCCUUUUAUUAUCAGUUUAGUUCGC--AUCCUUCGCCUUUUGGGCGGGAGUUAUGUGUGGCAACUGAUA--UUUUCCCAGGCUCCUGAGGGA ..(((..(.....)..(((((.(((((..........((((((((..((((((--((.(((((((.....))).))))..))))).)))))))))))--.))))))))))......))). ( -37.20) >DroYak_CAF1 28515 97 - 1 -------------------UGUGGGAAUCCCUUUUAUUAUCAGUUUAGUUCGC--AUUCUUCGCCUUUUGGGCUCGAGUUCUGUGUGGCAACUGAUA--UUUUCCCAGGCUCCCGUGGGA -------------------..((((((..........((((((((..((.(((--((.(((.(((.....)))..)))....)))))))))))))))--..))))))...(((....))) ( -26.80) >consensus UACCCAAGAGCCUCAUGCCGGUGGGAAUCCCUUUUAUUAUCAGUUUAGUUCGC__CUCCUUCGCCUUUUGGGCGGGAGUUAUGUGUGGCAACUGAUA__UUUUCCCAGGCUCCCGGGGGA ..(((..(((((.........((((((..........((((((((..((((((..((((..((((.....))))))))....))).)))))))))))....)))))))))))....))). (-27.98 = -30.30 + 2.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:13 2006