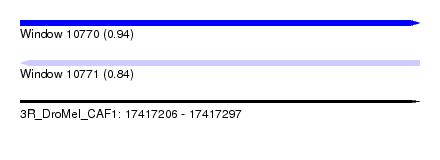
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,417,206 – 17,417,297 |
| Length | 91 |
| Max. P | 0.935969 |

| Location | 17,417,206 – 17,417,297 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -20.21 |
| Energy contribution | -21.10 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
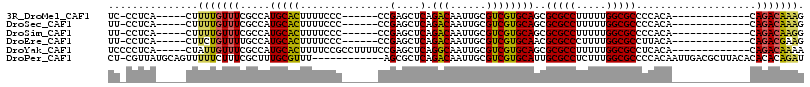
>3R_DroMel_CAF1 17417206 91 + 27905053 CUUUGUCUG-------------UGUGGGGCGCCAAAAAGGCGCGCUGCACGACGCAAUUGUCUGAGCUCGG------GGGAAAAGUGCAUGGCGAAACAAAAG-----UGAGG-GA .((((((((-------------((..(.(((((.....))))).)..))))..(((.((.((((....)))------).))....)))..)))))).......-----.....-.. ( -32.30) >DroSec_CAF1 21363 91 + 1 CUUUGUCUG-------------UGUGGGGCGCCAAAAAGGCGCGCUGCACGACGCAAUUGUCUGAGCUCGG------GGGAAAAGUGCAUGGCGAAACAAAAG-----UGAGG-AA .((((((((-------------((..(.(((((.....))))).)..))))..(((.((.((((....)))------).))....)))..)))))).......-----.....-.. ( -32.30) >DroSim_CAF1 18647 91 + 1 CCUUGUCUG-------------UGUGGGGCGCCAAAAAGGCGCGCUGCACGACGCAAUUGUCUGAGCUCGG------GGGAAAAGUGCAUGGCGAAACAAAAG-----UGAGG-AA (((((((.(-------------((..(.(((((.....))))).)..))))))(((.((.((((....)))------).))....)))...............-----.))))-.. ( -34.40) >DroEre_CAF1 20270 91 + 1 CUUCGUCUG-------------UGUAAGGCGCCAAAAGGGCGCGUUGCACGACGCAAUUGUCUGAGCUCGG------GGGAAAAGUGCAUGGCAAAACAGAAG-----UGAGG-AA (((((((.(-------------(((((.(((((.....))))).)))))))))(((.((.((((....)))------).))....)))...........))))-----.....-.. ( -33.60) >DroYak_CAF1 18413 98 + 1 UUUUGUCUG-------------UGUGAGGCGCCAAAAAGGCGCGCUGCACGACGCAAUUGCCUGAGCUCGGAAAAGGCGGAAAAGUGCAUGGCGAAACAAUAG-----UGAGGGGA (((((((((-------------(((.(((((((.....))))).)))))))..(((.((((((...........)))))).....)))..)))))))......-----........ ( -29.60) >DroPer_CAF1 27462 103 + 1 AUCUGUGUGUGUAAGCGUCAAUUGUGGGGCGCCAAAGAGGCGCAAUGCACGACGCAAUUGUCUGAGCGCU------------AAACGCAAAGCGAAAGAAAAACUGCAUAACG-AG ...(((.((((((.(((((...((((..(((((.....)))))..)))).))))).....(((...((((------------........))))..))).....)))))))))-.. ( -33.70) >consensus CUUUGUCUG_____________UGUGGGGCGCCAAAAAGGCGCGCUGCACGACGCAAUUGUCUGAGCUCGG______GGGAAAAGUGCAUGGCGAAACAAAAG_____UGAGG_AA ......................(((((.(((((.....))))).)))))...(((...(((((...(((........)))...)).)))..)))...................... (-20.21 = -21.10 + 0.89)


| Location | 17,417,206 – 17,417,297 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.72 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17417206 91 - 27905053 UC-CCUCA-----CUUUUGUUUCGCCAUGCACUUUUCCC------CCGAGCUCAGACAAUUGCGUCGUGCAGCGCGCCUUUUUGGCGCCCCACA-------------CAGACAAAG ..-.....-----..(((((((.((..(((((..(((..------..)))....(((......))))))))))(((((.....)))))......-------------.))))))). ( -23.70) >DroSec_CAF1 21363 91 - 1 UU-CCUCA-----CUUUUGUUUCGCCAUGCACUUUUCCC------CCGAGCUCAGACAAUUGCGUCGUGCAGCGCGCCUUUUUGGCGCCCCACA-------------CAGACAAAG ..-.....-----..(((((((.((..(((((..(((..------..)))....(((......))))))))))(((((.....)))))......-------------.))))))). ( -23.70) >DroSim_CAF1 18647 91 - 1 UU-CCUCA-----CUUUUGUUUCGCCAUGCACUUUUCCC------CCGAGCUCAGACAAUUGCGUCGUGCAGCGCGCCUUUUUGGCGCCCCACA-------------CAGACAAGG ..-.....-----..(((((((.((..(((((..(((..------..)))....(((......))))))))))(((((.....)))))......-------------.))))))). ( -23.40) >DroEre_CAF1 20270 91 - 1 UU-CCUCA-----CUUCUGUUUUGCCAUGCACUUUUCCC------CCGAGCUCAGACAAUUGCGUCGUGCAACGCGCCCUUUUGGCGCCUUACA-------------CAGACGAAG ..-.....-----(.(((((.......(((((..(((..------..)))....(((......))))))))..(((((.....))))).....)-------------)))).)... ( -21.80) >DroYak_CAF1 18413 98 - 1 UCCCCUCA-----CUAUUGUUUCGCCAUGCACUUUUCCGCCUUUUCCGAGCUCAGGCAAUUGCGUCGUGCAGCGCGCCUUUUUGGCGCCUCACA-------------CAGACAAAA ........-----...((((((.((..(((((.....(((..((.((.......)).))..)))..)))))))(((((.....)))))......-------------.)))))).. ( -25.40) >DroPer_CAF1 27462 103 - 1 CU-CGUUAUGCAGUUUUUCUUUCGCUUUGCGUUU------------AGCGCUCAGACAAUUGCGUCGUGCAUUGCGCCUCUUUGGCGCCCCACAAUUGACGCUUACACACACAGAU ..-((((((((((((..(((..((((........------------))))...))).)))))))..(((....(((((.....)))))..)))...)))))............... ( -26.40) >consensus UU_CCUCA_____CUUUUGUUUCGCCAUGCACUUUUCCC______CCGAGCUCAGACAAUUGCGUCGUGCAGCGCGCCUUUUUGGCGCCCCACA_____________CAGACAAAG ...............(((((((.....(((((...............(....).(((......))))))))..(((((.....)))))....................))))))). (-18.82 = -18.72 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:58 2006