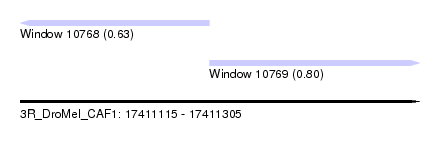
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,411,115 – 17,411,305 |
| Length | 190 |
| Max. P | 0.800931 |

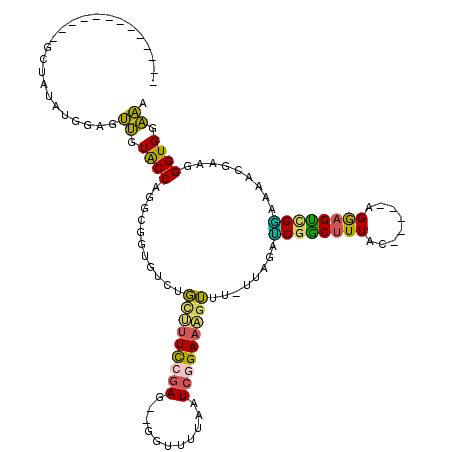
| Location | 17,411,115 – 17,411,205 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.69 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
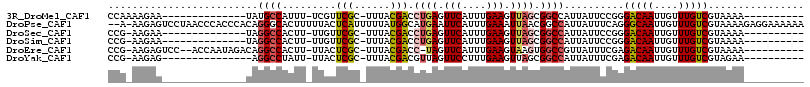
>3R_DroMel_CAF1 17411115 90 - 27905053 CCAAAAGAA--------------UAUGCCAUUU-UCGUUCGC-UUUACGACCUGAGUUCAUUUGAAGUUAGCGGCCAUUAUUCCGGGACAAUUGUUUGUCGUAAAA---------- ((....(((--------------((.(((....-((((....-...)))).((((.(((....))).)))).)))...))))).))(((((....)))))......---------- ( -20.30) >DroPse_CAF1 17135 113 - 1 --A-AAGAGUCCUAACCCACCCACAGGGCACUUUUUACUCAUUUUUAUGGCAUGAAUUCAUUUGAAAUUAACGGCCAUUAUUUCAGGGCAAUUGUUUGUCGUAAAAGAGGAAAAAA --.-.....((((..(((.......)))...(((((((........(((((.(((.(((....))).)))...)))))........(((((....))))))))))))))))..... ( -23.20) >DroSec_CAF1 15202 89 - 1 CCG-AAGAA--------------UAGGCCACUU-UUGUUCGC-UUUACGACCUGAGUUCAUUUGAAGUUAGCGGCCAUUAUUCCGGGACAAUUGUUUGUCGUAAAA---------- (((-..(((--------------((((((....-....(((.-....))).((((.(((....))).)))).))))..))))))))(((((....)))))......---------- ( -24.30) >DroSim_CAF1 12497 89 - 1 CCG-AAGAA--------------UAGGCCACUU-UUGUUCGC-UUUACGACCUGAGUUCAUUUGAAGUUAGCGGCCAUUAUUCCGGGACAAUUGUUUGUCGUAAAA---------- (((-..(((--------------((((((....-....(((.-....))).((((.(((....))).)))).))))..))))))))(((((....)))))......---------- ( -24.30) >DroEre_CAF1 13946 100 - 1 CCG-AAGAGUCC--ACCAAUAGACAGGCCACUU-UUACUCGC-UUUACGACC-UAGUUCAUUUGAAGUAAGUGGCCGUUAUUUCGAGACAAUUGUUUGUCGUAAAA---------- .((-(((.(((.--.......))).(((((((.-....(((.-....)))..-((.(((....))).)))))))))....))))).(((((....)))))......---------- ( -26.20) >DroYak_CAF1 12203 88 - 1 CCG-AAGAG---------------AGGCCUAUU-UUACUCGC-UUUACGACGUUAGUUCCUUUGAAGUUAGCGGCCAUUAUUUCGAGACAAUUGUUUGUCGUAGAA---------- .((-(((.(---------------(((((....-....(((.-....))).((((((((....))).))))))))).)).))))).(((((....)))))......---------- ( -21.00) >consensus CCG_AAGAA______________UAGGCCACUU_UUACUCGC_UUUACGACCUGAGUUCAUUUGAAGUUAGCGGCCAUUAUUCCGGGACAAUUGUUUGUCGUAAAA__________ .........................((((.........(((......))).((((.(((....))).)))).))))..........(((((....)))))................ (-14.48 = -14.70 + 0.22)
| Location | 17,411,205 – 17,411,305 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.33 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -18.22 |
| Energy contribution | -16.75 |
| Covariance contribution | -1.47 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
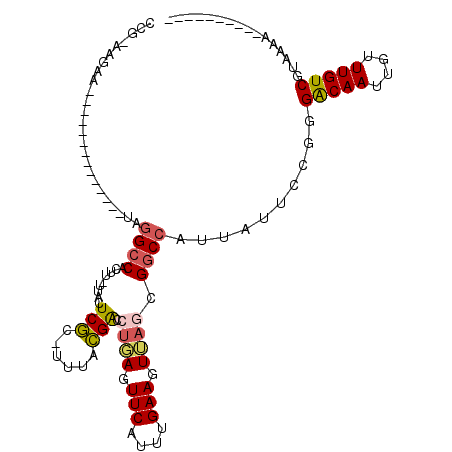
>3R_DroMel_CAF1 17411205 100 + 27905053 -------------GGUAUAUGGAGUUGUACCAGGCGGUAUCUGUUUUCCGAG--GCUUUUAAUCGGAAAGUUUUUUAUAUCGGCUUUAC-----AGGAGUUGAAAAACGAAGGGUGGAAA -------------((((((......)))))).(.((((((...((((((((.--........))))))))......)))))).)(((((-----....(((....))).....))))).. ( -25.20) >DroPse_CAF1 17248 96 + 1 ---------------------GCGCCGUGCCAGGCGCUGUUCCCGAUUCGAAGUGGU-UUUAUAGGAGCUGCG-GUAGCGCGGCAUUACUUGAAAGAAGUCGUUAAAUAAAUGGUGGGG- ---------------------(((((......)))))....((((.(((.(((((((-.........((((((-....)))))))))))))))).....(((((.....))))))))).- ( -29.70) >DroSec_CAF1 15291 98 + 1 -------------GCUAUAUAGAGUUGUACCAGGCGGUGUCUGCUUUCCGAG--GGU-UUAAUCGGAAAGUUU-UUAGAUCGACUUUAC-----AGGAGUCGGAAAACGGAGGGUGGAAA -------------...........((.((((...(.(((((((((((((((.--...-....)))))))))..-..))))((((((...-----..))))))....)).)..)))).)). ( -28.00) >DroSim_CAF1 12586 99 + 1 -------------GCUAUAUAGAGUUGUACCAGGCGGUGUCUGCUUUCCGAG--GGUUUUAAUCGGAAAGUUU-UUAGACCGACUUUAC-----AGGAGUCGGAAAACGGAGGGUGGAAA -------------...........((.((((...(.((.((((((((((((.--........)))))))))..-..)))(((((((...-----..)))))))...)).)..)))).)). ( -29.70) >DroEre_CAF1 14046 112 + 1 GACUCCAGCAAACGCUAUAUGGAGUUGUACCAGGCUGUGCCAGUUUUCCGAG--GGUUUUCAUCAGAAAGUUU-UUAAACCGGCUUUAC-----AGGAGCUGGAAAACGAAGGGUGGAAA (((((((((....))....))))))).((((.(((...))).((((((((((--..(((((....)))))..)-)).....(((((...-----..))))))))))))....)))).... ( -34.00) >DroYak_CAF1 12291 107 + 1 GAUUCC-----ACGCUAUAUGGAGUUGUACCAGGCGAUGCCAGUUUUCCGAA--GGUUUUCAUCAGAAUGUUU-UUAGAUCUGCUUUAC-----AGGAGCUGGAAAACGUAGGGUGAAAA .....(-----((.((((.(((.(((((.....))))).)))((((((((.(--(((((.(((....)))...-..))))))((((...-----..)))))))))))))))).))).... ( -29.20) >consensus _____________GCUAUAUGGAGUUGUACCAGGCGGUGUCUGCUUUCCGAG__GGUUUUAAUCGGAAAGUUU_UUAGAUCGGCUUUAC_____AGGAGUCGGAAAACGAAGGGUGGAAA ........................((.((((...........(((((((((...........)))))))))........((((((((........)))))))).........)))).)). (-18.22 = -16.75 + -1.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:56 2006