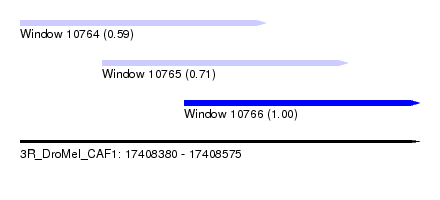
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,408,380 – 17,408,575 |
| Length | 195 |
| Max. P | 0.998632 |

| Location | 17,408,380 – 17,408,500 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17408380 120 + 27905053 ACGUAUCAUACACUUAAGAUGACUAAGCCAACUUAGUAGAACCGACUAAACCCUCAAAACUUUUGCCGCUUCAAAAACGGUUAGGUCUUGAACUAAAGAUGUGCCAAGCUGAAGUGGAAA .((..((...(((....).))((((((....)))))).))..)).....................((((((((.....(((.(.(((((......))))).))))....))))))))... ( -23.70) >DroSec_CAF1 12423 118 + 1 ACGUAUGAUACACUUAAGAUGACUAAGUCAACUUAGUAGAACCGCCUAAACCCUCAAAACUUUAGCCACUUCAAAAACGGUUAAGUCUUGAACUAAAGAU--GCCAAGCUGAAGUGGAAA .....(((.....(((((.((((...)))).)))))(((......))).....))).........((((((((.....(((...(((((......)))))--)))....))))))))... ( -22.90) >DroSim_CAF1 9712 118 + 1 ACGUAUGAUACACUUAAGAUGACUAAGUCAACUUAGUUGAACCGCCUAAACCCUCAAAACUUUAGCCACUUCAAAAACGGUUAGGUCUUGAACUAAAGAU--GCCAAGCUGAAGUGGAAA ..........((.(((((.((((...)))).))))).))..........................((((((((.....(((...(((((......)))))--)))....))))))))... ( -24.00) >DroEre_CAF1 11198 118 + 1 AACUAUGUUACACUUAAGAUGACUAAGUCCACUUAGUAGACCCGCCUAAACCCACAAAACUUUAGCCACUUCAAAAACGAUUGGGUCUUGAACUAAAGAU--GCCAAGCUGAAGUGGAAA .....................((((((....))))))............................((((((((.......(((((((((......)))))--.))))..))))))))... ( -23.50) >DroYak_CAF1 9515 118 + 1 AUAUAUGUUACGUUCAAGAUGACUAAGUCCACUUAGUAGAACCGCAUAAGCCCACAAAACUUUAGCCACUUGAAAAACGUUUAGGUCUUGAACUAAAGAU--GCCAAGCUAAAGUGGAAA ...(((((...((((......((((((....)))))).)))).)))))..........((((((((..((((((.....))))))((((......)))).--.....))))))))..... ( -23.50) >consensus ACGUAUGAUACACUUAAGAUGACUAAGUCAACUUAGUAGAACCGCCUAAACCCUCAAAACUUUAGCCACUUCAAAAACGGUUAGGUCUUGAACUAAAGAU__GCCAAGCUGAAGUGGAAA .....................((((((....))))))............................((((((((.....(((...(((((......)))))..)))....))))))))... (-20.44 = -20.88 + 0.44)


| Location | 17,408,420 – 17,408,540 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.19 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -25.38 |
| Energy contribution | -26.42 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17408420 120 + 27905053 ACCGACUAAACCCUCAAAACUUUUGCCGCUUCAAAAACGGUUAGGUCUUGAACUAAAGAUGUGCCAAGCUGAAGUGGAAACACGGCGGCUGAUGAUGACGCGAUAAGGGGAAAGGAAAGG ...........(((.....((((((.(((.(((.....(((.(.(((((......))))).)))).(((((..(((....)))..))))).....))).))).))))))...)))..... ( -35.40) >DroSec_CAF1 12463 118 + 1 ACCGCCUAAACCCUCAAAACUUUAGCCACUUCAAAAACGGUUAAGUCUUGAACUAAAGAU--GCCAAGCUGAAGUGGAAACACGGUGGCUGAUGAUGACGCGAUAAGGGGAAAAGAAAGG .((.(((...((.(((.....(((((((((.......(((((..(((((......)))))--....)))))..(((....))))))))))))...))).).)...))))).......... ( -32.50) >DroSim_CAF1 9752 118 + 1 ACCGCCUAAACCCUCAAAACUUUAGCCACUUCAAAAACGGUUAGGUCUUGAACUAAAGAU--GCCAAGCUGAAGUGGAAACACGGCGGCUGAUGAUGACGCGAUAAGGGGAAAGGAAAGG .((.(((...((((.......((((((.((.......(((((.((((((......)))..--))).)))))..(((....))))).))))))((......))...))))...)))...)) ( -32.80) >DroEre_CAF1 11238 118 + 1 CCCGCCUAAACCCACAAAACUUUAGCCACUUCAAAAACGAUUGGGUCUUGAACUAAAGAU--GCCAAGCUGAAGUGGAAACACGGCGGCUGAUGAUGACGCGAUAAGGGGAAAGGAAAGG .((.(((...(((........((((((.............(((((((((......)))))--.))))(((...(((....))))))))))))((......))....)))...)))...)) ( -32.70) >DroYak_CAF1 9555 118 + 1 ACCGCAUAAGCCCACAAAACUUUAGCCACUUGAAAAACGUUUAGGUCUUGAACUAAAGAU--GCCAAGCUAAAGUGGAAACACGGCGGCUGAUGAUGACGCGAUAAGGGGAAAGGAAAGG .((.(((.((((..(.....((((((..((((((.....))))))((((......)))).--.....))))))(((....))))..)))).)))....(.(.....).)....))..... ( -25.40) >consensus ACCGCCUAAACCCUCAAAACUUUAGCCACUUCAAAAACGGUUAGGUCUUGAACUAAAGAU__GCCAAGCUGAAGUGGAAACACGGCGGCUGAUGAUGACGCGAUAAGGGGAAAGGAAAGG .((.(((...(((........((((((...........(((...(((((......)))))..)))..(((...(((....))))))))))))((......))....)))...)))...)) (-25.38 = -26.42 + 1.04)

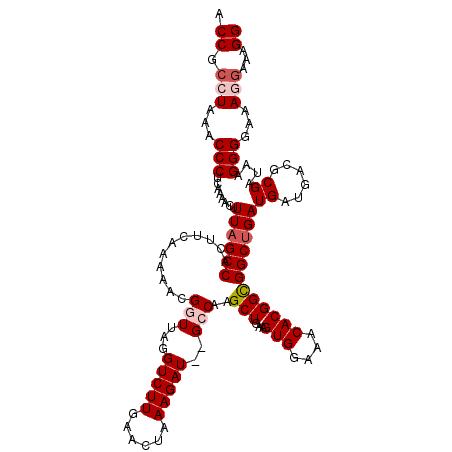
| Location | 17,408,460 – 17,408,575 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -31.18 |
| Energy contribution | -31.26 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17408460 115 + 27905053 UUAGGUCUUGAACUAAAGAUGUGCCAAGCUGAAGUGGAAACACGGCGGCUGAUGAUGACGCGAUAAGGGGAAAGGAAAGGGCCAGGCCAAAGCGAAACUGGCUCGGCUCGGCUGG ..(.(((((......))))).)(((.(((((..(((....)))..)))))............................(((((..((((.........))))..))))))))... ( -39.20) >DroSec_CAF1 12503 108 + 1 UUAAGUCUUGAACUAAAGAU--GCCAAGCUGAAGUGGAAACACGGUGGCUGAUGAUGACGCGAUAAGGGGAAAAGAAAGGGCCAGGCCAAAGCGAAACUGGCUC-----GGCUGG ....(((((......)))))--(((.(((((..(((....)))..)))))............................(((((((............)))))))-----)))... ( -32.80) >DroSim_CAF1 9792 108 + 1 UUAGGUCUUGAACUAAAGAU--GCCAAGCUGAAGUGGAAACACGGCGGCUGAUGAUGACGCGAUAAGGGGAAAGGAAAGGGCCAGGCCAAAGCGAAACUGGCUC-----GGCUGG ....(((((......)))))--(((.(((((..(((....)))..)))))............................(((((((............)))))))-----)))... ( -34.90) >DroEre_CAF1 11278 108 + 1 UUGGGUCUUGAACUAAAGAU--GCCAAGCUGAAGUGGAAACACGGCGGCUGAUGAUGACGCGAUAAGGGGAAAGGAAAGGGCCAGGCCAAAGCGAAACUGGCUC-----GGCUGG ....(((((......)))))--(((.(((((..(((....)))..)))))............................(((((((............)))))))-----)))... ( -34.90) >DroYak_CAF1 9595 108 + 1 UUAGGUCUUGAACUAAAGAU--GCCAAGCUAAAGUGGAAACACGGCGGCUGAUGAUGACGCGAUAAGGGGAAAGGAAAGGGCCAGGCCAAAGCGAAACUGGCUC-----GGCUGG ....(((((......)))))--(((.((((...(((....)))...))))............................(((((((............)))))))-----)))... ( -31.40) >consensus UUAGGUCUUGAACUAAAGAU__GCCAAGCUGAAGUGGAAACACGGCGGCUGAUGAUGACGCGAUAAGGGGAAAGGAAAGGGCCAGGCCAAAGCGAAACUGGCUC_____GGCUGG ....(((((......)))))..(((.(((((..(((....)))..)))))............................(((((((............))))))).....)))... (-31.18 = -31.26 + 0.08)

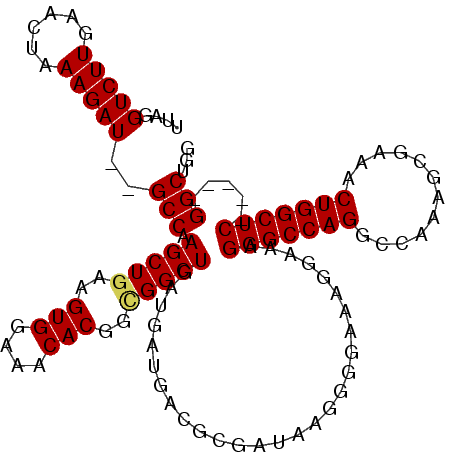

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:53 2006