
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,430,709 – 2,430,849 |
| Length | 140 |
| Max. P | 0.688645 |

| Location | 2,430,709 – 2,430,829 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -36.95 |
| Consensus MFE | -30.21 |
| Energy contribution | -31.13 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
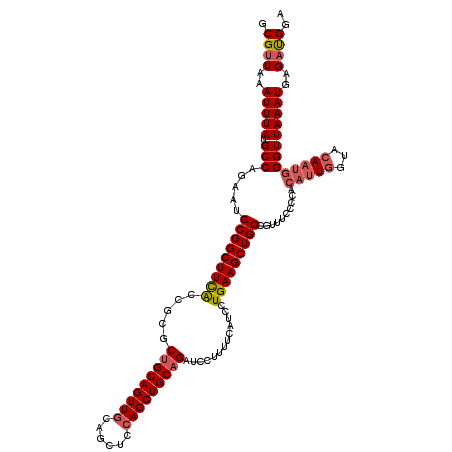
>3R_DroMel_CAF1 2430709 120 + 27905053 GCGUUAAAUUUAUGCCAGAAUGCGGCUUCAACGCGCCGCAGUUGCAGCUCCAGCUGCAGAUCCUUUUCAUCCUGAAGCUGCCGUUUCCCCACAUUUGUACAAUGGGUUAAAUGAGAUGGA ((((.......)))).....((((((........)))))).(((((((....))))))).(((.((((((...(((((....)))))((((...........))))....)))))).))) ( -36.50) >DroSec_CAF1 143447 111 + 1 GCGUUAAAUUUAUGCCAGAAUGCGGCUUCACCGCGCUGCAGUUGCAGCUCCAGCUGCAGAUCCUUUUCAUCCUGAAGCUGCCGUU---------UGGUACAAUGGGUUAAAUGAGAUGGA .((((.(((((((((((((..(((((((((.....(((((((((......))))))))).............)))))))))..))---------))))...))))))).))))....... ( -36.87) >DroSim_CAF1 137630 120 + 1 GCGUUAAAUUUAUGCCAGAAUGCGGCUUCACCGCGCUGCAGUUGCAGCUCCAGCUGCAGAUCCUUUUCAUCCUGAAGCUGCCGUUUCCCCACAUUGGUACAAUGGGUUAAAUGAGAUGGA ..............(((....(((((((((.....(((((((((......))))))))).............)))))))))(((((..((.(((((...)))))))..)))))...))). ( -36.57) >DroEre_CAF1 139255 120 + 1 GCGUUAAAUUUAUGCCAGAAUGCGGCUUCGCCGCGCUGCAGUUGCAACUCGAGCUGCAGAUCCUUUUAAUCCUGAAGCUGCCGUUUUCCCACAUUGGUACAAUGGGUUAAAUGAGACGGA ((((.......))))(((...(((((...))))).((((((((........))))))))............)))...(((((((((..((.(((((...)))))))..))))).).))). ( -35.90) >DroYak_CAF1 133455 120 + 1 GCGUUAAAUUUAUGCCAGAAUGCGGCUUUACCUCGCUGCAGUUGCAGCUCCAGCUGCAGAUCCUUUUCAUCCUGAAGCUGCCGUUUCCCCACAUUGGUACAAUGGGUUAAAUGAGCUGCA ((((.......)))).....((((((........))))))..(((((((((((((.(((............))).)))))..((((..((.(((((...)))))))..)))))))))))) ( -38.90) >consensus GCGUUAAAUUUAUGCCAGAAUGCGGCUUCACCGCGCUGCAGUUGCAGCUCCAGCUGCAGAUCCUUUUCAUCCUGAAGCUGCCGUUUCCCCACAUUGGUACAAUGGGUUAAAUGAGAUGGA .((((..(((((.(((.....(((((((((.....(((((((((......))))))))).............)))))))))..........(((((...)))))))))))))..)))).. (-30.21 = -31.13 + 0.92)

| Location | 2,430,709 – 2,430,829 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -35.76 |
| Consensus MFE | -30.08 |
| Energy contribution | -31.00 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2430709 120 - 27905053 UCCAUCUCAUUUAACCCAUUGUACAAAUGUGGGGAAACGGCAGCUUCAGGAUGAAAAGGAUCUGCAGCUGGAGCUGCAACUGCGGCGCGUUGAAGCCGCAUUCUGGCAUAAAUUUAACGC .(((..........(((((.........)))))(((...((((((((((..((...((...)).)).))))))))))...((((((........)))))))))))).............. ( -37.20) >DroSec_CAF1 143447 111 - 1 UCCAUCUCAUUUAACCCAUUGUACCA---------AACGGCAGCUUCAGGAUGAAAAGGAUCUGCAGCUGGAGCUGCAACUGCAGCGCGGUGAAGCCGCAUUCUGGCAUAAAUUUAACGC ..........................---------..((((((.(((..........))).)))).((..(((((((....)))))((((.....))))..))..))..........)). ( -31.10) >DroSim_CAF1 137630 120 - 1 UCCAUCUCAUUUAACCCAUUGUACCAAUGUGGGGAAACGGCAGCUUCAGGAUGAAAAGGAUCUGCAGCUGGAGCUGCAACUGCAGCGCGGUGAAGCCGCAUUCUGGCAUAAAUUUAACGC (((...((((((..(((((.........)))))(((.(....).))).))))))...)))......((..(((((((....)))))((((.....))))..))..))............. ( -37.80) >DroEre_CAF1 139255 120 - 1 UCCGUCUCAUUUAACCCAUUGUACCAAUGUGGGAAAACGGCAGCUUCAGGAUUAAAAGGAUCUGCAGCUCGAGUUGCAACUGCAGCGCGGCGAAGCCGCAUUCUGGCAUAAAUUUAACGC ..(((...(((((.(((((.........)))))......((.(((.(((((((.....))))((((((....)))))).))).)))(((((...)))))......)).)))))...))). ( -35.10) >DroYak_CAF1 133455 120 - 1 UGCAGCUCAUUUAACCCAUUGUACCAAUGUGGGGAAACGGCAGCUUCAGGAUGAAAAGGAUCUGCAGCUGGAGCUGCAACUGCAGCGAGGUAAAGCCGCAUUCUGGCAUAAAUUUAACGC ((((((((......(((((.........))))).......(((((.((((.(....)...)))).)))))))))))))......(((.((.....))((......))..........))) ( -37.60) >consensus UCCAUCUCAUUUAACCCAUUGUACCAAUGUGGGGAAACGGCAGCUUCAGGAUGAAAAGGAUCUGCAGCUGGAGCUGCAACUGCAGCGCGGUGAAGCCGCAUUCUGGCAUAAAUUUAACGC ..............(((((.........)))))....((((((.(((..........))).)))).((..(((((((....)))))(((((...)))))..))..))..........)). (-30.08 = -31.00 + 0.92)



| Location | 2,430,749 – 2,430,849 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -21.38 |
| Energy contribution | -22.46 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
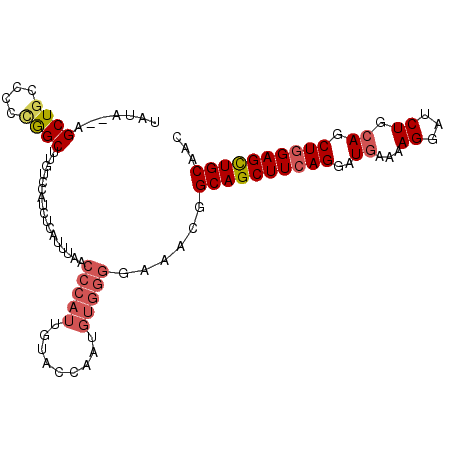
>3R_DroMel_CAF1 2430749 100 - 27905053 UAUA--AGCUGCACCCAGCUUGUCCAUCUCAUUUAACCCAUUGUACAAAUGUGGGGAAACGGCAGCUUCAGGAUGAAAAGGAUCUGCAGCUGGAGCUGCAAC .(((--(((((....)))))))).............(((((.........)))))......((((((((((..((...((...)).)).))))))))))... ( -31.70) >DroSec_CAF1 143487 91 - 1 UAUA--AGCUGCCCCUGGCUUGUCCAUCUCAUUUAACCCAUUGUACCA---------AACGGCAGCUUCAGGAUGAAAAGGAUCUGCAGCUGGAGCUGCAAC .(((--((((......))))))).........................---------....((((((((((..((...((...)).)).))))))))))... ( -23.50) >DroSim_CAF1 137670 100 - 1 UAUA--AGCUGCCCCCGGCUUGUCCAUCUCAUUUAACCCAUUGUACCAAUGUGGGGAAACGGCAGCUUCAGGAUGAAAAGGAUCUGCAGCUGGAGCUGCAAC .(((--(((((....)))))))).............(((((((...))))).))(....).((((((((((..((...((...)).)).))))))))))... ( -31.30) >DroEre_CAF1 139295 102 - 1 CACACCAGCUUAUCCCGGCUUGUCCGUCUCAUUUAACCCAUUGUACCAAUGUGGGAAAACGGCAGCUUCAGGAUUAAAAGGAUCUGCAGCUCGAGUUGCAAC ....((.....((((.((((.(.((((.....((..(((((((...))))).))..)))))))))))...)))).....))...((((((....)))))).. ( -25.50) >DroYak_CAF1 133495 100 - 1 UAUA--AGCUGGCCCCAGCGUCUGCAGCUCAUUUAACCCAUUGUACCAAUGUGGGGAAACGGCAGCUUCAGGAUGAAAAGGAUCUGCAGCUGGAGCUGCAAC ....--.((((....))))...((((((((......(((((.........))))).......(((((.((((.(....)...)))).))))))))))))).. ( -38.30) >consensus UAUA__AGCUGCCCCCGGCUUGUCCAUCUCAUUUAACCCAUUGUACCAAUGUGGGGAAACGGCAGCUUCAGGAUGAAAAGGAUCUGCAGCUGGAGCUGCAAC .......((((....)))).................(((((.........)))))......((((((((((..((...((...)).)).))))))))))... (-21.38 = -22.46 + 1.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:39 2006