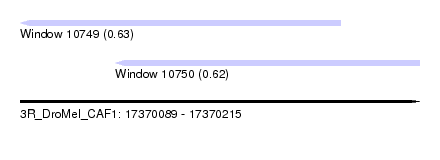
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,370,089 – 17,370,215 |
| Length | 126 |
| Max. P | 0.628724 |

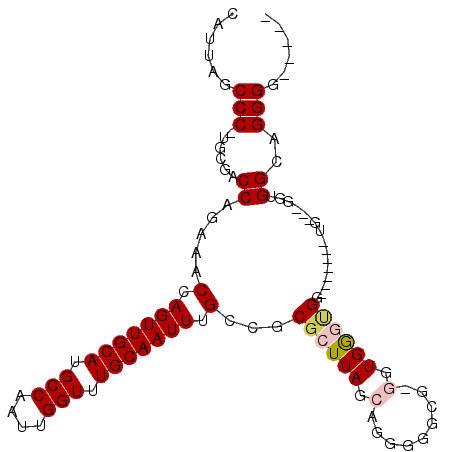
| Location | 17,370,089 – 17,370,190 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -37.33 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.45 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
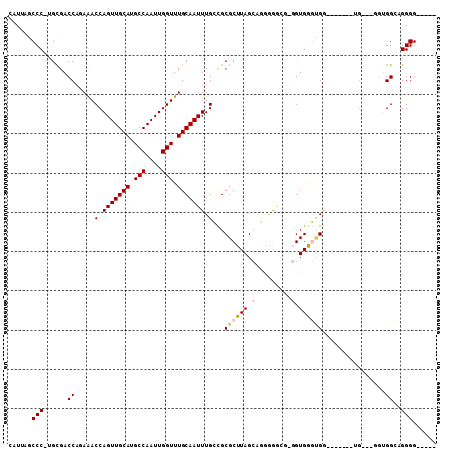
>3R_DroMel_CAF1 17370089 101 - 27905053 CAUUAGCCC-UGUGACCAGAAACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGCCGCGCUUAGCAGGAGGAUGGGUGGGUGG-------UGCCUAGUGGCCGGGGUAAAU ..(((.(((-.((.((.((..((((((((((.(((....))).))))))).(((.((((((.........)))))))))))-------)..)).)).)).))).))).. ( -36.50) >DroPse_CAF1 23503 89 - 1 CAUUAGCCCGCGCGACCA-AGACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGGCGCGCUUAGAACGGAACG--GUGAUUGA-------UG-----AGGAAGGGG----- ......((((((((.(((-(.....((((((.(((....))).)))))))))))))))((((.((......--))..))))-------..-----.....))).----- ( -28.60) >DroEre_CAF1 21411 99 - 1 CAUUAGCCC-UGUGACCAGAAACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGCCGCGCUUAGCAGGGGGCGGGGUGGGCGG-------UGCCUGGUGGCCGGGG--AGU ......(((-.((.(((((..((((((((((.(((....))).))))))).(((.((((..((.....))..)))))))))-------)..))))).)).))).--... ( -46.20) >DroYak_CAF1 28730 107 - 1 CAUUAGCCC-UGUGACCAGAAACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGCCGCGCUUAGCAGGGGGCGGGGUGGGUGGUGCAUGGUGCCUGGUGGCCGGGGU-AGU .((((.(((-.((.(((((..((((((((((.(((....))).))))(((..((.(((((......))))).))..)))...)).))))..))))).)).))).)-))) ( -49.00) >DroAna_CAF1 15665 88 - 1 AAUUAUCCC-AGCGACC-GAAACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGGUGCGCUUAGCCGAAG----GGUGGGAGG-------U----GGUGGGAGGGGU---- ....(((((-..(.(((-...((((((((((.(((....))).))))).)))))((.(((.(((....----)))..))))-------)----))).)..)))))---- ( -35.10) >DroPer_CAF1 16037 89 - 1 CAUUAGCCCGCGCGACCA-AGACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGGCGCGCUUAGAACGGAACG--GUGAUUGA-------UG-----AGGAAGGGG----- ......((((((((.(((-(.....((((((.(((....))).)))))))))))))))((((.((......--))..))))-------..-----.....))).----- ( -28.60) >consensus CAUUAGCCC_UGCGACCAGAAACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGCCGCGCUUAGCAGGGGGCG_GGUGGGUGG_______UG___GGUGGCAGGGG_____ ......(((......((.....(.(((((((.(((....))).))))))).)...((((((.(.........).))))))................))..)))...... (-16.95 = -17.45 + 0.50)

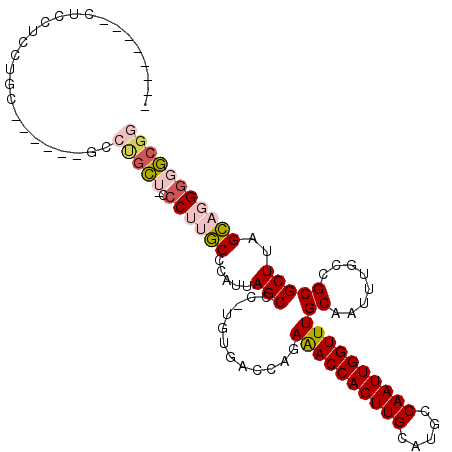
| Location | 17,370,119 – 17,370,215 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.54 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -20.79 |
| Energy contribution | -21.35 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17370119 96 - 27905053 --------CUCCUCCUGU------GCCUGCU-UCCUUGCCCAUUAGCCC-UGUGACCAGAAACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGCCGCGCUUAGCAGGAGGAUG --------.(((((((((------(((.((.-...((((.........(-((....)))((((((((((.....))))))))))))))...)).).))...))))))))).. ( -33.60) >DroPse_CAF1 23522 96 - 1 --------UCCAUUCCGC------ACCCGUACCCCCAUCCCAUUAGCCCGCGCGACCA-AGACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGGCGCGCUUAGAACGGAACG- --------(((.(((.((------....))...................(((((.(((-(.....((((((.(((....))).)))))))))))))))...))).)))...- ( -27.70) >DroSec_CAF1 18557 96 - 1 --------CUCCUCCUGC------GCCUGCU-UCCUUGCCCAUUAGCCC-UGUGACCAGAAACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGCCGCGCUUAGCAGGGGGCGG --------(.((((((((------(((.((.-...((((.........(-((....)))((((((((((.....))))))))))))))...)).).))...)))))))).). ( -32.80) >DroSim_CAF1 27171 96 - 1 --------CUCCUCCUGC------GCCUGCU-UCCUUGCCCAUUAGCCC-UGUGACCAGAAACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGCCGCGCUUAGCAGGGGGCGG --------(.((((((((------(((.((.-...((((.........(-((....)))((((((((((.....))))))))))))))...)).).))...)))))))).). ( -32.80) >DroEre_CAF1 21439 110 - 1 ACCUCCUCCUCGUCCUGCCUCUCAUCCUGCU-CCCUUGCCCAUUAGCCC-UGUGACCAGAAACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGCCGCGCUUAGCAGGGGGCGG ..........................((((.-(((((((.....(((.(-.(..(...(((((((((((.....)))))))))).)...)..).).)))..))))))))))) ( -31.70) >DroPer_CAF1 16056 96 - 1 --------UCCAUUCCGC------ACCCGUACCCCCAUCCCAUUAGCCCGCGCGACCA-AGACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGGCGCGCUUAGAACGGAACG- --------(((.(((.((------....))...................(((((.(((-(.....((((((.(((....))).)))))))))))))))...))).)))...- ( -27.70) >consensus ________CUCCUCCUGC______GCCUGCU_CCCUUGCCCAUUAGCCC_UGUGACCAGAAACCAGUUGCAUGCCAAUUGGUUUGCAAUUUGCCGCGCUUAGCAGGGGGCGG ..........................(((((..((((((.....(((............((((((((((.....))))))))))((........)))))..))))))))))) (-20.79 = -21.35 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:37 2006