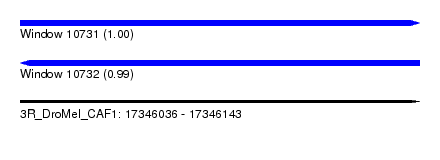
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,346,036 – 17,346,143 |
| Length | 107 |
| Max. P | 0.996743 |

| Location | 17,346,036 – 17,346,143 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -34.74 |
| Consensus MFE | -23.19 |
| Energy contribution | -23.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
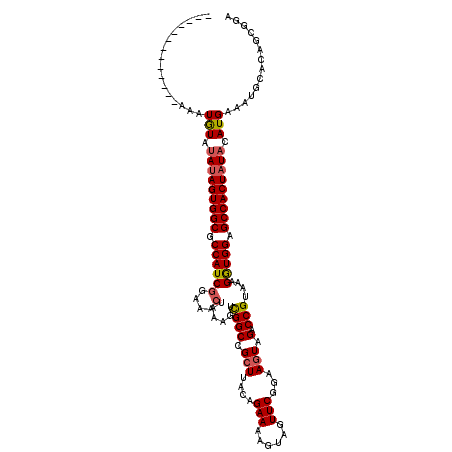
>3R_DroMel_CAF1 17346036 107 + 27905053 -------------AAAUGUAUAUAGUGGCGCCAUCGGAAACUAAAGUCGGCCGCUACCAGAAUAGUAGUUCGGAAGUAGACCGUAAAGGUGGAGCCACUAUACAUGAAAUGCUCAGCGGA -------------..(((((..(((((((.((((((....)......((((..((.((.((((....)))))).))..).)))....))))).))))))))))))............... ( -35.20) >DroSim_CAF1 4035 107 + 1 -------------AAAUGUAUAUAGUGGCGCCAUCGGAAACCAAAGUCGGCCGCUUACAGAAAAGUAGUUCAGAAGUAGACCGUAAAGGUGGAGCCACUAUAUAUGAAAUGCACAGCGGA -------------....((((((((((((.((((((....)......((((..(((...(((......)))..)))..).)))....))))).))))))))))))............... ( -34.80) >DroYak_CAF1 5439 119 + 1 GAGUAACGGGUAAAAAUAUACAGAGUGGCGCCAUCGGAAAAUAAAGUUGGCCGCUUAAAGAAAAUUAGUUCGGUAAUAGACCGUAAAGAUGGAGCCACUAUACAUGAAUUGCCCAG-GCA ..((...((((((..........((((((.(((((((..(((...)))..))..................((((.....))))....))))).)))))).........))))))..-)). ( -34.21) >consensus _____________AAAUGUAUAUAGUGGCGCCAUCGGAAACUAAAGUCGGCCGCUUACAGAAAAGUAGUUCGGAAGUAGACCGUAAAGGUGGAGCCACUAUACAUGAAAUGCACAGCGGA ................(((.(((((((((.((((((....)......((((.(((....(((......)))...))).).)))....))))).))))))))).))).............. (-23.19 = -23.87 + 0.67)

| Location | 17,346,036 – 17,346,143 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -29.61 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.37 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
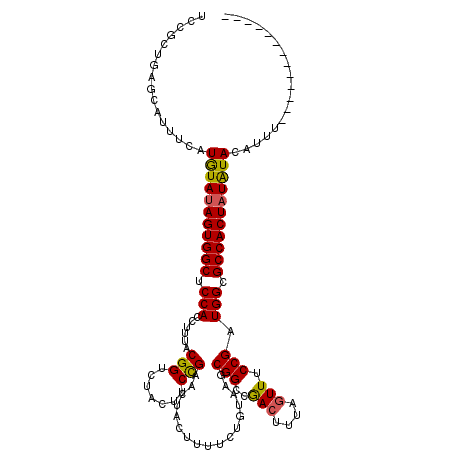
>3R_DroMel_CAF1 17346036 107 - 27905053 UCCGCUGAGCAUUUCAUGUAUAGUGGCUCCACCUUUACGGUCUACUUCCGAACUACUAUUCUGGUAGCGGCCGACUUUAGUUUCCGAUGGCGCCACUAUAUACAUUU------------- ................(((((((((((.(((......(((.......)))..(((((.....)))))(((..(((....))).))).))).))))))))))).....------------- ( -30.20) >DroSim_CAF1 4035 107 - 1 UCCGCUGUGCAUUUCAUAUAUAGUGGCUCCACCUUUACGGUCUACUUCUGAACUACUUUUCUGUAAGCGGCCGACUUUGGUUUCCGAUGGCGCCACUAUAUACAUUU------------- ................(((((((((((.(((......(((((..((((.(((......))).).))).)))))...((((...))))))).))))))))))).....------------- ( -29.30) >DroYak_CAF1 5439 119 - 1 UGC-CUGGGCAAUUCAUGUAUAGUGGCUCCAUCUUUACGGUCUAUUACCGAACUAAUUUUCUUUAAGCGGCCAACUUUAUUUUCCGAUGGCGCCACUCUGUAUAUUUUUACCCGUUACUC ...-..(((.((...((((((((((((.(((((....((((.....))))..................((.............))))))).))))))..))))))..)).)))....... ( -29.32) >consensus UCCGCUGAGCAUUUCAUGUAUAGUGGCUCCACCUUUACGGUCUACUUCCGAACUACUUUUCUGUAAGCGGCCGACUUUAGUUUCCGAUGGCGCCACUAUAUACAUUU_____________ ................(((((((((((.(((......(((.......))).................(((..(((....))).))).))).))))))))))).................. (-23.58 = -23.37 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:21 2006