
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,345,320 – 17,345,475 |
| Length | 155 |
| Max. P | 0.737834 |

| Location | 17,345,320 – 17,345,435 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -29.79 |
| Consensus MFE | -25.79 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17345320 115 + 27905053 UACCAAAUCGAUU-UUCAC----CAUCCCCAAACUGGGGGCUUCAACCCGUUUUAACGGCUGCCAUAAUCGCACCCAGCCGCAAACUGACGGGAAAAAGUUGAUUGAUGAUAAUGACCCA .....((((((((-((...----..(((((.....)))))......((((((....((((((.............))))))......))))))..))))))))))............... ( -32.42) >DroSim_CAF1 3334 116 + 1 UUCCAAAUCGAUUUUUCAC----CAUCCCCAAACUGUGGGCUUCAACCCGUUUUAACUGCUGCCAUAAUCGCACCCAGCCGCAGACUGACGGGAAAACAUUGAUUGAUGAUAAUGACCCA .((..((((((((((((..----....((((.....)))).......(((((....((((.((..............)).))))...)))))))))).)))))))...)).......... ( -25.24) >DroYak_CAF1 4163 120 + 1 UUCCAAAUCGAUUUUUCACUCCCAAUCCCCAAACUGGGGACUUCAACCCGUUUUAACGGCUGCCAUAAUCGCACCCAGCCGCAGACUGACGGGAAAGCAUUGAUUGAUGAUAAUGACCCA .((..((((((((((((........(((((.....))))).......(((((....((((((.............))))))......)))))))))).)))))))...)).......... ( -31.72) >consensus UUCCAAAUCGAUUUUUCAC____CAUCCCCAAACUGGGGGCUUCAACCCGUUUUAACGGCUGCCAUAAUCGCACCCAGCCGCAGACUGACGGGAAAACAUUGAUUGAUGAUAAUGACCCA .....(((((((.............(((((.....)))))......((((((....((((((.............))))))......)))))).....)))))))............... (-25.79 = -26.02 + 0.23)

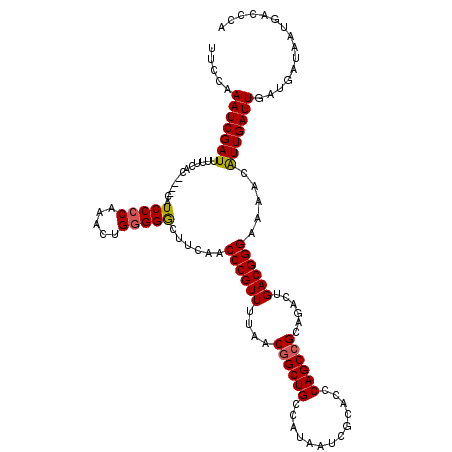

| Location | 17,345,355 – 17,345,475 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -24.55 |
| Energy contribution | -26.55 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17345355 120 + 27905053 CUUCAACCCGUUUUAACGGCUGCCAUAAUCGCACCCAGCCGCAAACUGACGGGAAAAAGUUGAUUGAUGAUAAUGACCCAAAUUCGUAAAUAAACGAUAGCAUCGUUAUUAUCAUUGCAU ......((((((....((((((.............))))))......)))))).....((....((((((((((((.......((((......)))).....))))))))))))..)).. ( -33.32) >DroSim_CAF1 3370 120 + 1 CUUCAACCCGUUUUAACUGCUGCCAUAAUCGCACCCAGCCGCAGACUGACGGGAAAACAUUGAUUGAUGAUAAUGACCCAAAUUCGUAAAUAAACGAUAGAAUCGCUUUUAUCAUUGCUU ......((((((....((((.((..............)).))))...))))))............((((((((..........((((......))))...........)))))))).... ( -25.04) >DroYak_CAF1 4203 117 + 1 CUUCAACCCGUUUUAACGGCUGCCAUAAUCGCACCCAGCCGCAGACUGACGGGAAAGCAUUGAUUGAUGAUAAUGACCCAAAUUCGUAAAUAAACGAUAGCAUCGUUAUU---AUAGCUA ......((((((....((((((.............))))))......))))))..(((........((((((((((.......((((......)))).....))))))))---)).))). ( -28.42) >consensus CUUCAACCCGUUUUAACGGCUGCCAUAAUCGCACCCAGCCGCAGACUGACGGGAAAACAUUGAUUGAUGAUAAUGACCCAAAUUCGUAAAUAAACGAUAGCAUCGUUAUUAUCAUUGCUU ......((((((....((((((.............))))))......))))))...........((((((((((((.......((((......)))).....))))))))))))...... (-24.55 = -26.55 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:19 2006