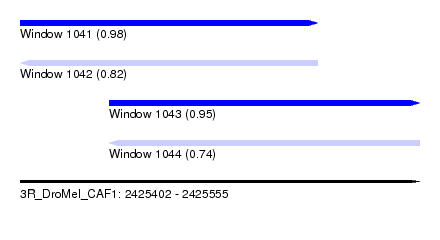
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,425,402 – 2,425,555 |
| Length | 153 |
| Max. P | 0.977987 |

| Location | 2,425,402 – 2,425,516 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.01 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -14.15 |
| Energy contribution | -16.27 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2425402 114 + 27905053 AAC---GCU---UUGCGUGUUGCUCUUCUGAUGUCGUUUAACCAUUAUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGAC ...---(((---..(((.(((((((..........((((((.......((((((......))))))......))))))(((((.....)))))))).)))).))).)))........... ( -34.52) >DroSec_CAF1 138373 114 + 1 AAC---GCU---UUGCGUGUUGCUCUUCUGAUGCCGUUUAACCAUUAUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGAA ...---(((---..(((.(((((((..........((((((.......((((((......))))))......))))))(((((.....)))))))).)))).))).)))........... ( -34.52) >DroSim_CAF1 132348 114 + 1 AAC---GCU---UUGCGUGUUGCUCUUCUGAUGCCGUUUAACCAUUAUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGAA ...---(((---..(((.(((((((..........((((((.......((((((......))))))......))))))(((((.....)))))))).)))).))).)))........... ( -34.52) >DroEre_CAF1 133437 114 + 1 AAU---GUU---UUGCGUUUUGCUCUUCUGAUGUCGUUUAACCAUUAUGUGGCAUGAGUUUGCCAUUUUCAUUUAAAUGGUUCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUUGCGAU ...---...---(((((....((.......((((((..((.....))..))))))(((...(((((((......))))))).)))....(((((((....))))))))).....))))). ( -29.10) >DroYak_CAF1 127567 87 + 1 AAA---GUU---UUGCG-----------UGAUGUCGUUUAGCCAUUAUGUGGCAUGAAUUUGCCACUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGCCAACUCU---------------- ..(---(((---..(((-----------(((((.(.....).))))))((((((......))))))...........((((((.....)))))).)).))))..---------------- ( -25.20) >DroAna_CAF1 134005 98 + 1 UAUUCAGCUUUAUUGCGUAUGACUUUUCUGAUGUCGGUUAACCACUUUUUG-AGUAAAAGUGUCAUUUCCAUUUUAC---UCCAUUUGGUAAUGACAGAAAU------------------ ((((((((......))....(((.........)))((....))......))-))))....(((((((.(((......---......))).))))))).....------------------ ( -19.00) >consensus AAC___GCU___UUGCGUGUUGCUCUUCUGAUGUCGUUUAACCAUUAUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGA_ ......((......))..................(((((((.......((((((......))))))......)))))))........(.(((((((....))))))).)........... (-14.15 = -16.27 + 2.12)

| Location | 2,425,402 – 2,425,516 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.01 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -9.77 |
| Energy contribution | -11.22 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2425402 114 - 27905053 GUCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAUAAUGGUUAAACGACAUCAGAAGAGCAACACGCAA---AGC---GUU ((((......(((((..(((....)))(((((.....))))).............(((((......))))).....)))))...))))..............((((..---.))---)). ( -27.70) >DroSec_CAF1 138373 114 - 1 UUCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAUAAUGGUUAAACGGCAUCAGAAGAGCAACACGCAA---AGC---GUU ..((((.....(((...(((....)))(((((.....))))).(((((.......(((((......)))))........))))).))).........((.....))..---)))---).. ( -28.86) >DroSim_CAF1 132348 114 - 1 UUCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAUAAUGGUUAAACGGCAUCAGAAGAGCAACACGCAA---AGC---GUU ..((((.....(((...(((....)))(((((.....))))).(((((.......(((((......)))))........))))).))).........((.....))..---)))---).. ( -28.86) >DroEre_CAF1 133437 114 - 1 AUCGCAAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGAACCAUUUAAAUGAAAAUGGCAAACUCAUGCCACAUAAUGGUUAAACGACAUCAGAAGAGCAAAACGCAA---AAC---AUU .((((......))....((((((.(((........)))(((((((...(((....(((((......)))))))))))))))...)))).)).))...((.....))..---...---... ( -21.20) >DroYak_CAF1 127567 87 - 1 ----------------AGAGUUGGCUCGCUGCCAAGAGCAGCAUUUAAAUGAAAGUGGCAAAUUCAUGCCACAUAAUGGCUAAACGACAUCA-----------CGCAA---AAC---UUU ----------------....((((((.(((((.....)))))............((((((......)))))).....)))))).........-----------.....---...---... ( -23.50) >DroAna_CAF1 134005 98 - 1 ------------------AUUUCUGUCAUUACCAAAUGGA---GUAAAAUGGAAAUGACACUUUUACU-CAAAAAGUGGUUAACCGACAUCAGAAAAGUCAUACGCAAUAAAGCUGAAUA ------------------.((((((((...((((....((---((((((((.......)).)))))))-)......)))).....)))...)))))........((......))...... ( -17.70) >consensus _UCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAUAAUGGUUAAACGACAUCAGAAGAGCAACACGCAA___AGC___GUU ...................((((....(((((.....))))).............(((((......))))).............))))................................ ( -9.77 = -11.22 + 1.45)


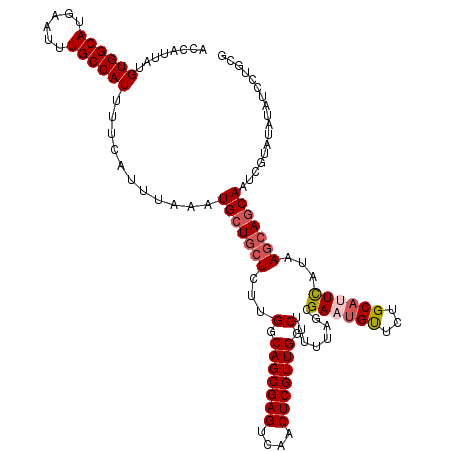
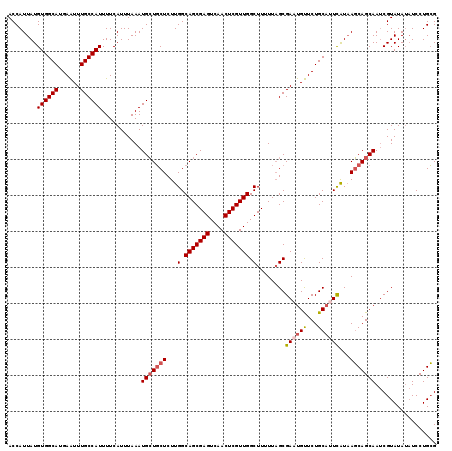
| Location | 2,425,436 – 2,425,555 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -29.15 |
| Energy contribution | -30.28 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2425436 119 + 27905053 ACCAUUAUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGACUGUUCUGCAUUCAUAAGCAGCAAUCGUAUAUAUCCUGCG ........((((((......))))))...........(((((((..((((((((((....)))))))(((....))).............)))..)))))))..((((.......)))) ( -34.30) >DroSec_CAF1 138407 119 + 1 ACCAUUAUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGAAUGUUCUGCAUUCAUAAGCAGCAAUCGUAUAUAUCCUGCG ........((((((......))))))...........(((((((.....(((((((....)))))))(((....)))(((((.....)))))...)))))))..((((.......)))) ( -36.30) >DroSim_CAF1 132382 119 + 1 ACCAUUAUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGAAUGUUCUGCAUUCAUAAGCAGCAAUCGUAUAUAUCCUGCG ........((((((......))))))...........(((((((.....(((((((....)))))))(((....)))(((((.....)))))...)))))))..((((.......)))) ( -36.30) >DroEre_CAF1 133471 118 + 1 ACCAUUAUGUGGCAUGAGUUUGCCAUUUUCAUUUAAAUGGUUCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUUGCGAUUGCUCUGCCUUUGUUACCAGCAA-CGUAUAAUUCCUGGG .(((......(((..(((...(((((((......))))))).)))....(((((((....))))))))))....((((.(((((..((....))....)))))-)))).......))). ( -32.10) >consensus ACCAUUAUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGAAUGUUCUGCAUUCAUAAGCAGCAAUCGUAUAUAUCCUGCG ........((((((......))))))...........(((((((...(.(((((((....))))))).)........((((((...))))))...)))))))................. (-29.15 = -30.28 + 1.13)

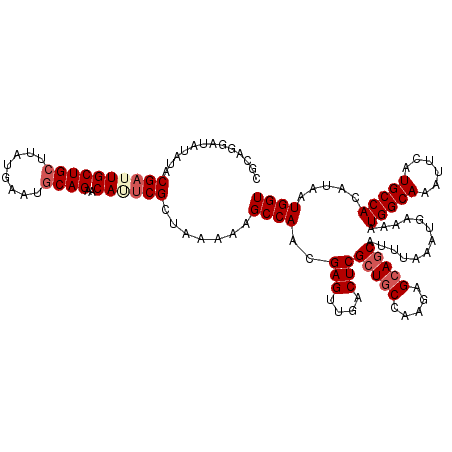
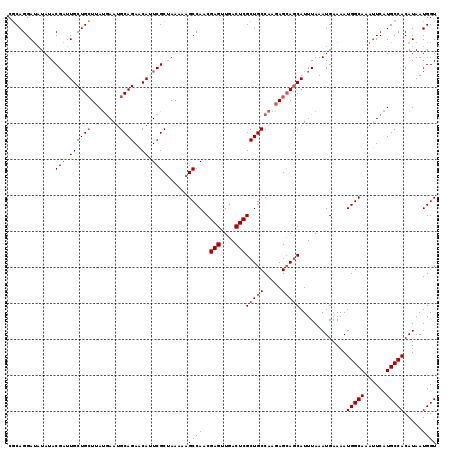
| Location | 2,425,436 – 2,425,555 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -24.19 |
| Energy contribution | -25.62 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2425436 119 - 27905053 CGCAGGAUAUAUACGAUUGCUGCUUAUGAAUGCAGAACAGUCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAUAAUGGU .(((((((.....((((((((((........))))..)))))).......((((..(((....)))(((((.....))))).............))))..)))).)))........... ( -33.60) >DroSec_CAF1 138407 119 - 1 CGCAGGAUAUAUACGAUUGCUGCUUAUGAAUGCAGAACAUUCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAUAAUGGU .(((((((.....(((.((((((........))))..)).))).......((((..(((....)))(((((.....))))).............))))..)))).)))........... ( -30.40) >DroSim_CAF1 132382 119 - 1 CGCAGGAUAUAUACGAUUGCUGCUUAUGAAUGCAGAACAUUCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAUAAUGGU .(((((((.....(((.((((((........))))..)).))).......((((..(((....)))(((((.....))))).............))))..)))).)))........... ( -30.40) >DroEre_CAF1 133471 118 - 1 CCCAGGAAUUAUACG-UUGCUGGUAACAAAGGCAGAGCAAUCGCAAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGAACCAUUUAAAUGAAAAUGGCAAACUCAUGCCACAUAAUGGU .((((.(((.....)-)).)))).......(((((.((....))...........((((....)))))))))......((((((...(((....(((((......)))))))))))))) ( -31.10) >consensus CGCAGGAUAUAUACGAUUGCUGCUUAUGAAUGCAGAACAUUCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAUAAUGGU .............((((((((((........))))..)))))).......((((..(((....)))(((((.....))))).............(((((......))))).....)))) (-24.19 = -25.62 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:35 2006