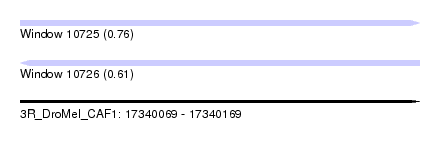
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,340,069 – 17,340,169 |
| Length | 100 |
| Max. P | 0.762586 |

| Location | 17,340,069 – 17,340,169 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 96.47 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17340069 100 + 27905053 UUGGAAGUGACCAGGGGGCGGAUGGGCGUGUGUCAGUCAAAAAAGUGGAUGCGAGUAUGAAAAAUACAAACGAACAGCCCGAACAGAAUGCGGUUGGUCA .......(((((...((((...((..((((((((...(......)..)))))..((((.....))))..)))..)))))).(((........)))))))) ( -24.40) >DroSec_CAF1 16027 100 + 1 UUGGAAGUGACCAGGGGGCGGAUGGGCGUGUGUCAGUCAAAAAAGUGGAUGCGAGUAUGAAAAAUACAAACGAACAGCCCAAACAGAAUGCGGUUGGUCA .......(((((...((((...((..((((((((...(......)..)))))..((((.....))))..)))..)))))).(((........)))))))) ( -24.30) >DroSim_CAF1 21623 100 + 1 UUGGAAGUGACCAGGGGGCGGAUGGGCGUGUGUCAGUCAAAAGAGUGGAUGCGAGUAUGAAAAAUACAAACGAACAGCCCGAACAGGAUGCGGUUGGUCA .......(((((...((((...((..(((((((((.((....)).).)))))..((((.....))))..)))..)))))).(((........)))))))) ( -25.80) >DroEre_CAF1 21309 100 + 1 UUGGAAGUGACCAGGGGGCCGAUGGGCGUGUGUCAGUCAAAAAAGUGGAUGCGAGUAUGAAAAAUACAAACGAACAGCCCGAACAGAAUGCGGUUGGUCA .......((((((....((((.(((((.((((((...(......)..)))))).((((.....)))).........))))).........)))))))))) ( -24.80) >DroYak_CAF1 19878 100 + 1 UUGGAAGUGACCAGGGGGCCGAUGGGCGUGUGUCAGUCAAAAAAGUGGAUGCGAGUAUGAAAAAUACAAACGAACAGCCCGAACAGAAUGCGGUUGGUCA .......((((((....((((.(((((.((((((...(......)..)))))).((((.....)))).........))))).........)))))))))) ( -24.80) >DroAna_CAF1 21874 100 + 1 UUGGAAGUGACCGGGGGGUGACCGGGCGAGUGUCAGUCAAAAAAGUGGAUGCGAGUAUGAAAAAUACAAACGAACAGCCCGAACAGAAUGCGGUUGGUCA ...((..((((((.(((....))((((..(((((...(......)..)))))..((((.....)))).........))))........).)))))).)). ( -27.60) >consensus UUGGAAGUGACCAGGGGGCGGAUGGGCGUGUGUCAGUCAAAAAAGUGGAUGCGAGUAUGAAAAAUACAAACGAACAGCCCGAACAGAAUGCGGUUGGUCA .......(((((...((((...((..((((((((...(......)..)))))..((((.....))))..)))..)))))).(((........)))))))) (-23.10 = -23.30 + 0.20)

| Location | 17,340,069 – 17,340,169 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 96.47 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -16.82 |
| Energy contribution | -17.15 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17340069 100 - 27905053 UGACCAACCGCAUUCUGUUCGGGCUGUUCGUUUGUAUUUUUCAUACUCGCAUCCACUUUUUUGACUGACACACGCCCAUCCGCCCCCUGGUCACUUCCAA ((((((...((.....))..(((((((((((..((((.....))))..))...((......))...)).))).))))..........))))))....... ( -18.40) >DroSec_CAF1 16027 100 - 1 UGACCAACCGCAUUCUGUUUGGGCUGUUCGUUUGUAUUUUUCAUACUCGCAUCCACUUUUUUGACUGACACACGCCCAUCCGCCCCCUGGUCACUUCCAA ((((((...((.....(..((((((((((((..((((.....))))..))...((......))...)).))).)))))..)))....))))))....... ( -20.30) >DroSim_CAF1 21623 100 - 1 UGACCAACCGCAUCCUGUUCGGGCUGUUCGUUUGUAUUUUUCAUACUCGCAUCCACUCUUUUGACUGACACACGCCCAUCCGCCCCCUGGUCACUUCCAA ((((((...((.....))..(((((((..((..((((.....))))..))...((.((....)).))..))).))))..........))))))....... ( -19.40) >DroEre_CAF1 21309 100 - 1 UGACCAACCGCAUUCUGUUCGGGCUGUUCGUUUGUAUUUUUCAUACUCGCAUCCACUUUUUUGACUGACACACGCCCAUCGGCCCCCUGGUCACUUCCAA ((((((...((.....))..((((((...((.(((.((..(((..................)))..)).))).))....))))))..))))))....... ( -20.67) >DroYak_CAF1 19878 100 - 1 UGACCAACCGCAUUCUGUUCGGGCUGUUCGUUUGUAUUUUUCAUACUCGCAUCCACUUUUUUGACUGACACACGCCCAUCGGCCCCCUGGUCACUUCCAA ((((((...((.....))..((((((...((.(((.((..(((..................)))..)).))).))....))))))..))))))....... ( -20.67) >DroAna_CAF1 21874 100 - 1 UGACCAACCGCAUUCUGUUCGGGCUGUUCGUUUGUAUUUUUCAUACUCGCAUCCACUUUUUUGACUGACACUCGCCCGGUCACCCCCCGGUCACUUCCAA (((((....((.....))..(((......((..((((.....))))..))...........((((((.........))))))...))))))))....... ( -18.60) >consensus UGACCAACCGCAUUCUGUUCGGGCUGUUCGUUUGUAUUUUUCAUACUCGCAUCCACUUUUUUGACUGACACACGCCCAUCCGCCCCCUGGUCACUUCCAA ((((((...((.....))..(((((((((((..((((.....))))..))...((......))...)).))).))))..........))))))....... (-16.82 = -17.15 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:15 2006