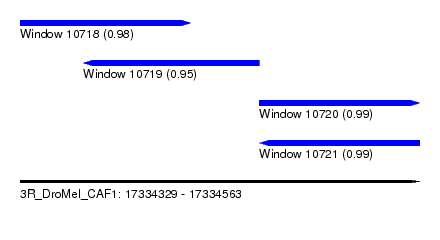
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,334,329 – 17,334,563 |
| Length | 234 |
| Max. P | 0.990777 |

| Location | 17,334,329 – 17,334,429 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -12.84 |
| Energy contribution | -13.57 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17334329 100 + 27905053 AGUGGCCGUCAUAAAGACGAUGGCAAAUGGCGGCUAAAUGGCCGAAGUUUCCCACUUU-CCCAGG-----AAAGGGAGCAGGGAAAGCGGCA-------AGUCGGGAAAGCAG .((.((((((........))))))......(((((.....((((...((((((...((-(((...-----...)))))..)))))).)))).-------))))).....)).. ( -36.50) >DroPse_CAF1 17816 87 + 1 ACCGACGAUCAUAAGGCUGAUGGCAAAUGGCGGCUAAAUGGGCAAAGUUUCCUACUUUUCUCAGGCCGGCAACGGGAACGGG------AACG-------G------------- .(((.(.((((......)))).)....((.(((((...((((.(((((.....))))).))))))))).)).)))...((..------..))-------.------------- ( -27.10) >DroSec_CAF1 10291 96 + 1 AGUGGCCGUCAUAAAGACGAUGGCAAAUGGCGGCUAAAUGACCGAAGUUUCCCACUUU-CCCAGG-----AAAGGGAGCAGGGAAAGCGGCA-------AGUCGGGAAA---- ....((((((........))))))......(((((...((.(((...((((((...((-(((...-----...)))))..)))))).)))))-------))))).....---- ( -33.90) >DroSim_CAF1 15899 96 + 1 AGUGGCCGUCAUAAAGACGAUGGCAAAUGGCGGCUAAAUGGCCGAAGUUUCCCACUUU-CCCAGG-----AAAGGGAGCAGGGAAAGCGGCA-------AGUCGGGAAA---- ....((((((........))))))......(((((.....((((...((((((...((-(((...-----...)))))..)))))).)))).-------))))).....---- ( -36.20) >DroYak_CAF1 13848 107 + 1 AGGGGCCGUCAUAAAGACGAUGGCAAAUGGCGGCUAAAUGGCCGAAGUUUCCCACUUU-CCCAGG-----AAAGGGAGCAGGAAAAGCGGGAUGCGAGAAGGCGGGAAAGUAG .(((((((((........))))))((((..(((((....)))))..)))))))(((((-(((...-----.......((.......)).....((......)))))))))).. ( -39.50) >DroPer_CAF1 18841 86 + 1 ACCGACGAUCAUAAGGCUGAUGGCAAAUGGCGGCUAAAUGGGCAAAGUUUCCAACUUUUCUCAGGCCGGCAACGGGAACG-C------AACG-------A------------- .(((.(.((((......)))).)....((.(((((...((((.(((((.....))))).))))))))).)).))).....-.------....-------.------------- ( -24.10) >consensus AGUGGCCGUCAUAAAGACGAUGGCAAAUGGCGGCUAAAUGGCCGAAGUUUCCCACUUU_CCCAGG_____AAAGGGAGCAGGGAAAGCGGCA_______AGUCGGGAAA____ ....((((((........)))))).......((((....))))...((((((.....................)))))).................................. (-12.84 = -13.57 + 0.72)
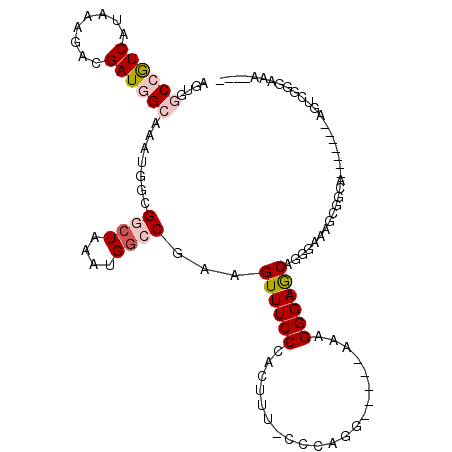
| Location | 17,334,366 – 17,334,469 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -22.30 |
| Energy contribution | -21.92 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17334366 103 - 27905053 AACGCGACUUAAGCGCCGUCCCGCCAGCUUUCCCGCCUUGCUGCUUUCCCGACU-------UGCCGCUUUCCCUGCUCCCUUUCCUGGGAAAGUGGGAAACUUCGGCCAU ...((.......))((((....((.(((...........))))).........(-------(.((((((((((.............)))))))))).))....))))... ( -29.72) >DroSec_CAF1 10328 87 - 1 AACGCGGCUUAAGCGCCCUCCCGCCCGC----------------UUUCCCGACU-------UGCCGCUUUCCCUGCUCCCUUUCCUGGGAAAGUGGGAAACUUCGGUCAU ...((((.....(((......)))))))----------------....((((.(-------(.((((((((((.............)))))))))).))...)))).... ( -29.82) >DroSim_CAF1 15936 87 - 1 AACGCGGCUUAAGCGCCCUCCCGCCCGC----------------UUUCCCGACU-------UGCCGCUUUCCCUGCUCCCUUUCCUGGGAAAGUGGGAAACUUCGGCCAU ...((((.....(((......)))))))----------------....((((.(-------(.((((((((((.............)))))))))).))...)))).... ( -29.62) >DroYak_CAF1 13885 110 - 1 AACGCGACUUAAGCGCCGUCCCGCCCGCUUUCCCGCCUUCCUACUUUCCCGCCUUCUCGCAUCCCGCUUUUCCUGCUCCCUUUCCUGGGAAAGUGGGAAACUUCGGCCAU ...((.......))((((.....(((((((((((................((......)).....((.......))..........)))))))))))......))))... ( -33.00) >consensus AACGCGACUUAAGCGCCCUCCCGCCCGC________________UUUCCCGACU_______UGCCGCUUUCCCUGCUCCCUUUCCUGGGAAAGUGGGAAACUUCGGCCAU ..(((.......))).......(((......................................((((((((((.............))))))))))((....)))))... (-22.30 = -21.92 + -0.38)

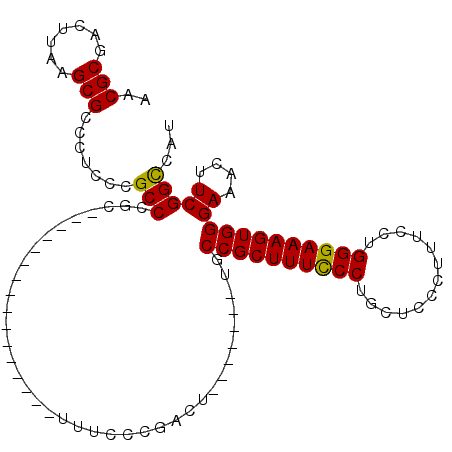
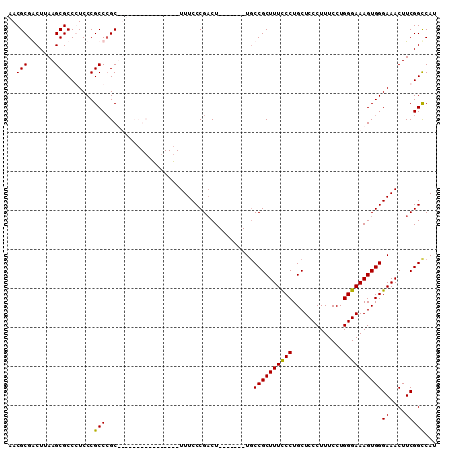
| Location | 17,334,469 – 17,334,563 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.65 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -16.78 |
| Energy contribution | -18.28 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17334469 94 + 27905053 AAUUGCCGCUUAGCAGGCGGCGUUUUC-AGGC----------AU-UUCAGACAUUUUUAGAGCCCAGUCCUCCU-------GGCUUAUCAGCCAUUUUGGGCCUAUUCCAGCU ....((((((.....))))))......-....----------..-............(((.((((((......(-------((((....)))))..)))))))))........ ( -27.60) >DroSec_CAF1 10415 75 + 1 AAUUGCCGCUUAGCAGGCGGCGUU-------------------------------UUUAGAGCCCAGUCAUCCU-------GGCUUAUCAGCCAUUUUGGGCCUAUUCCAACU ....((((((.....))))))(((-------------------------------..(((.((((((......(-------((((....)))))..)))))))))....))). ( -28.20) >DroSim_CAF1 16023 75 + 1 AAUUGCCGCUUAGCAGGCGGCGUU-------------------------------UUUAGAGCCCAGUCCUCCU-------GGCUUAUCAGCCAUUUUGGGCCUAUUCCAGCU ....((((((.....))))))(((-------------------------------..(((.((((((......(-------((((....)))))..)))))))))....))). ( -28.20) >DroEre_CAF1 15581 109 + 1 AAUUGCCGCUUAGCAGGCGGCCUUUUC-AGGCGUGUUCAGGCAU-UUCAGUCGUUUUUAGAGCCCAGUCCUCCUGG--ACUGGCUUAUCAGCCAUUUUGGGCCUAUUCCAGCU ....((((((.....))))))......-.(((.......(((..-....))).....(((.(((((((((....))--))(((((....)))))...)))))))).....))) ( -37.50) >DroYak_CAF1 13995 104 + 1 AAUUGCCGCUUAGCAGGCGGACUUUUC-AGGCGU-UUCAGUCGUUUUCAGACGUUUUCAGAGCCCAGUCCUCCU-------GGCUUAUCAGCCAUUUUGGGCCUAUUCCAGCU .....(((((.....))))).......-.(((..-....(((.......)))......((.((((((......(-------((((....)))))..))))))))......))) ( -28.10) >DroAna_CAF1 14441 89 + 1 AAUUGCCGCUUAGCAGGCCCAGUUGUCGAGGC----------A------A--------ACGGGGCAGUCCUCUCGGCAGUCGGCUUAUCAGCCAUUUUGGGCCUAUUCCAGCU .......(((....(((((((((((((((((.----------.------.--------((......))..)))))))))..((((....))))...)))))))).....))). ( -32.50) >consensus AAUUGCCGCUUAGCAGGCGGCGUUUUC_AGGC__________A______G_____UUUAGAGCCCAGUCCUCCU_______GGCUUAUCAGCCAUUUUGGGCCUAUUCCAGCU ....((((((.....))))))....................................(((.((((((....).........((((....))))....))))))))........ (-16.78 = -18.28 + 1.50)


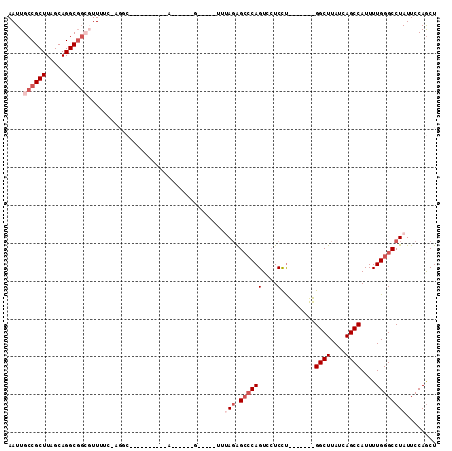
| Location | 17,334,469 – 17,334,563 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.65 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -17.33 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17334469 94 - 27905053 AGCUGGAAUAGGCCCAAAAUGGCUGAUAAGCC-------AGGAGGACUGGGCUCUAAAAAUGUCUGAA-AU----------GCCU-GAAAACGCCGCCUGCUAAGCGGCAAUU ........(((((((....(((((....))))-------)...))...((((.........))))...-..----------))))-).....(((((.......))))).... ( -27.40) >DroSec_CAF1 10415 75 - 1 AGUUGGAAUAGGCCCAAAAUGGCUGAUAAGCC-------AGGAUGACUGGGCUCUAAA-------------------------------AACGCCGCCUGCUAAGCGGCAAUU .(((....((((((((...(((((....))))-------).......))))).)))..-------------------------------)))(((((.......))))).... ( -28.20) >DroSim_CAF1 16023 75 - 1 AGCUGGAAUAGGCCCAAAAUGGCUGAUAAGCC-------AGGAGGACUGGGCUCUAAA-------------------------------AACGCCGCCUGCUAAGCGGCAAUU ........((((((((...(((((....))))-------).......))))).)))..-------------------------------...(((((.......))))).... ( -27.10) >DroEre_CAF1 15581 109 - 1 AGCUGGAAUAGGCCCAAAAUGGCUGAUAAGCCAGU--CCAGGAGGACUGGGCUCUAAAAACGACUGAA-AUGCCUGAACACGCCU-GAAAAGGCCGCCUGCUAAGCGGCAAUU ........((((((((...(((((....)))))((--((....))))))))).)))............-.((((.......((((-....)))).((.......))))))... ( -37.90) >DroYak_CAF1 13995 104 - 1 AGCUGGAAUAGGCCCAAAAUGGCUGAUAAGCC-------AGGAGGACUGGGCUCUGAAAACGUCUGAAAACGACUGAA-ACGCCU-GAAAAGUCCGCCUGCUAAGCGGCAAUU .((((.(.(((((......(((((....))))-------)...(((((((((........(((......)))......-..))))-....)))))))))).)...)))).... ( -30.89) >DroAna_CAF1 14441 89 - 1 AGCUGGAAUAGGCCCAAAAUGGCUGAUAAGCCGACUGCCGAGAGGACUGCCCCGU--------U------U----------GCCUCGACAACUGGGCCUGCUAAGCGGCAAUU .((((.(.((((((((...(((((....)))))..((.((((.((((......))--------)------)----------..)))).))..)))))))).)...)))).... ( -33.30) >consensus AGCUGGAAUAGGCCCAAAAUGGCUGAUAAGCC_______AGGAGGACUGGGCUCUAAA_____C______U__________GCCU_GAAAACGCCGCCUGCUAAGCGGCAAUU ..........(((((.....((((....)))).......((.....))))))).......................................(((((.......))))).... (-17.33 = -18.22 + 0.89)
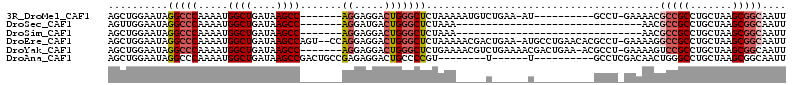
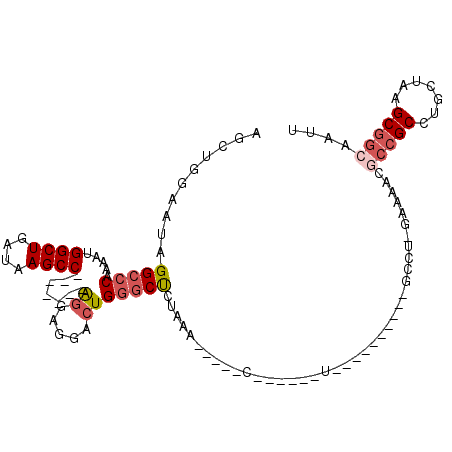
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:10 2006