
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,333,527 – 17,333,815 |
| Length | 288 |
| Max. P | 0.999395 |

| Location | 17,333,527 – 17,333,634 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -16.55 |
| Energy contribution | -16.22 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
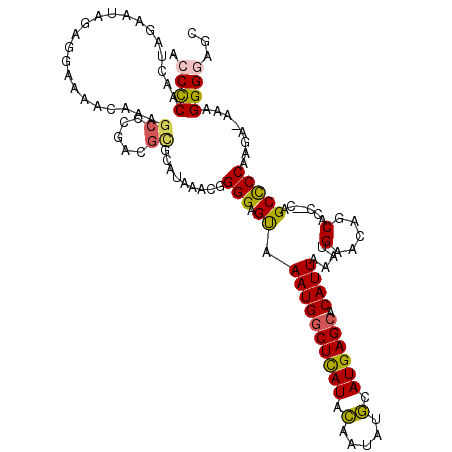
>3R_DroMel_CAF1 17333527 107 + 27905053 ---UU-------ACACGGCUAAUGCCCAGUA---CGGUGUAACUCAACUAGAUUAUAGGGCAUCAAAGCCCGACGCGCAUAAACGGGGAGCAAAUGGCUUAUACAAUAUGCAUGAGCACA ---((-------(((((((....)))(....---.)))))))((((...........((((......)))).....(((((......((((.....))))......))))).)))).... ( -28.50) >DroPse_CAF1 16940 118 + 1 AGAUGAGGUCUUGCAAA--CAAUGUUUUAUUUGUGGGUAUACCCCAACUAGAAAAGAGGAAUACAAAGCGUGAUGUACACGAGCGGGGAGUAAAUGGCUCAUUCAAUAUGCAUGAGCACA ........((((((...--.....(((((.(((.(((....)))))).)))))...............((((.....)))).))))))........((((((.(.....).))))))... ( -26.40) >DroSec_CAF1 9495 107 + 1 ---UU-------ACAUGACUAAAAUCAUGUU---CGUUGUAACCCAACUAGAUUAUAGGGCAUCAAAGCCCGACGCGCAUAAACGGGGAGCAAAUGGCUCAUACAAUAUGCAUGAGCACA ---..-------............((((((.---.((((((.(((............((((......)))).............)))((((.....)))).))))))..))))))..... ( -29.81) >DroSim_CAF1 15119 107 + 1 ---UU-------ACACGACUAAAAUCAUGUU---CGUUGUAACCCAACUAGAUUAUAGGGCAUCAAAGCCCGACGCGCAUAAACGGGGAGCAAAUAGCUCAUACAAUAUGCAUGAGCACA ---..-------............((((((.---.((((((.(((............((((......)))).............)))((((.....)))).))))))..))))))..... ( -29.91) >DroEre_CAF1 14706 106 + 1 ---UU-------GCAAGGCUAGUGCCUAGUU---UCGCAUA-CCCAACUAGACCAUAGGGCAUCAAAGCCCGGCGCGCAUAAACGGGGAGCAAAUGGCUCAUACAAUAUGCAUGAGCACA ---..-------.....(((.((((((((((---.......-...))))).......((((......)))))))))(((((......((((.....))))......)))))...)))... ( -32.40) >DroYak_CAF1 13043 107 + 1 ---UC-------ACCAGGCUAAUGCCUAGUC---CCGUAUAACCCAACUAGACCAUAGGGCAUCAAAGCCCGACGCGCAUAAACGGCGAGCAAAUGGCUCAUACAAUAUGCAUGGGCACA ---..-------...((((....)))).(((---((((...................((((......)))).....(((((......((((.....))))......)))))))))).)). ( -31.90) >consensus ___UU_______ACAAGGCUAAUGCCUAGUU___CGGUAUAACCCAACUAGAUUAUAGGGCAUCAAAGCCCGACGCGCAUAAACGGGGAGCAAAUGGCUCAUACAAUAUGCAUGAGCACA ............((.(((......))).))............(((............((((......))))..((........)))))........((((((.(.....).))))))... (-16.55 = -16.22 + -0.33)



| Location | 17,333,527 – 17,333,634 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -24.39 |
| Energy contribution | -23.57 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17333527 107 - 27905053 UGUGCUCAUGCAUAUUGUAUAAGCCAUUUGCUCCCCGUUUAUGCGCGUCGGGCUUUGAUGCCCUAUAAUCUAGUUGAGUUACACCG---UACUGGGCAUUAGCCGUGU-------AA--- ((((((((.....((((((...(((((..((.....))..))).))...((((......)))))))))).....)))).))))...---(((..(((....)))..))-------).--- ( -27.40) >DroPse_CAF1 16940 118 - 1 UGUGCUCAUGCAUAUUGAAUGAGCCAUUUACUCCCCGCUCGUGUACAUCACGCUUUGUAUUCCUCUUUUCUAGUUGGGGUAUACCCACAAAUAAAACAUUG--UUUGCAAGACCUCAUCU (((((....)))))....((((((............))))))(((((........)))))...........((.((((((.......(((((((....)))--))))....)))))).)) ( -22.30) >DroSec_CAF1 9495 107 - 1 UGUGCUCAUGCAUAUUGUAUGAGCCAUUUGCUCCCCGUUUAUGCGCGUCGGGCUUUGAUGCCCUAUAAUCUAGUUGGGUUACAACG---AACAUGAUUUUAGUCAUGU-------AA--- ((.((((((((.....)))))))))).(((..((((((....)))....((((......))))............)))...)))..---.(((((((....)))))))-------..--- ( -32.60) >DroSim_CAF1 15119 107 - 1 UGUGCUCAUGCAUAUUGUAUGAGCUAUUUGCUCCCCGUUUAUGCGCGUCGGGCUUUGAUGCCCUAUAAUCUAGUUGGGUUACAACG---AACAUGAUUUUAGUCGUGU-------AA--- ...((((((((.....)))))))).................((((((.(((((......))))((.((((..((((.....)))).---.....)))).))).)))))-------).--- ( -32.60) >DroEre_CAF1 14706 106 - 1 UGUGCUCAUGCAUAUUGUAUGAGCCAUUUGCUCCCCGUUUAUGCGCGCCGGGCUUUGAUGCCCUAUGGUCUAGUUGGG-UAUGCGA---AACUAGGCACUAGCCUUGC-------AA--- ((.((((((((.....))))))))))(((((..((((.(((.((.((..((((......))))..)))).))).))))-...))))---)...((((....))))...-------..--- ( -37.70) >DroYak_CAF1 13043 107 - 1 UGUGCCCAUGCAUAUUGUAUGAGCCAUUUGCUCGCCGUUUAUGCGCGUCGGGCUUUGAUGCCCUAUGGUCUAGUUGGGUUAUACGG---GACUAGGCAUUAGCCUGGU-------GA--- ...(((((.(((((.....(((((.....))))).....))))).(((.((((......)))).))).......))))).......---.(((((((....)))))))-------..--- ( -38.00) >consensus UGUGCUCAUGCAUAUUGUAUGAGCCAUUUGCUCCCCGUUUAUGCGCGUCGGGCUUUGAUGCCCUAUAAUCUAGUUGGGUUACACCG___AACAAGACAUUAGCCUUGU_______AA___ ((((((((((((((......((((.....))))......))))))....((((......))))...........)))).))))........((((((....))))))............. (-24.39 = -23.57 + -0.83)


| Location | 17,333,554 – 17,333,672 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.87 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -15.48 |
| Energy contribution | -15.12 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17333554 118 + 27905053 AACUCAACUAGAUUAUAGGGCAUCAAAGCCCGACGCGCAUAAACGGGGAGCAAAUGGCUUAUACAAUAUGCAUGAGCACAUUAAAUGAACAGCACC--CAGCCCCAAGAAAAAGGGGAGC .................((((......)))).....((.......(((.((.....((((((.(.....).)))))).(((...)))....)).))--)..((((........)))).)) ( -29.00) >DroPse_CAF1 16978 116 + 1 ACCCCAACUAGAAAAGAGGAAUACAAAGCGUGAUGUACACGAGCGGGGAGUAAAUGGCUCAUUCAAUAUGCAUGAGCACAUUAAAUGAAAAACUCG--CAGCUCCAAUG--AUGGGGAGC .(((((.......................(((.....)))((((.(.((((.((((((((((.(.....).)))))).))))...(....))))).--).)))).....--.)))))... ( -32.50) >DroSec_CAF1 9522 118 + 1 AACCCAACUAGAUUAUAGGGCAUCAAAGCCCGACGCGCAUAAACGGGGAGCAAAUGGCUCAUACAAUAUGCAUGAGCACAUUAAAUGAACAGCGCC--CAGCCCCAAGAAAAAGGGGAGC .................((((......))))...((((.......(....).((((((((((.(.....).)))))).)))).........)))).--...((((........))))... ( -32.00) >DroAna_CAF1 13612 107 + 1 ACCCCAACUAGACUGAAGUUU------------CUGGCUUAAACAGGGAGUAAAUGGCUUAUAAAAAAUUCAUGAGAACAUUAAAUGAGCAGCACUCGCAGCCCCAAA-GACAGGGGAGC .((((..(((((........)------------))))........(((.((.....((((((......(((....)))......)))))).((....)).)))))...-....))))... ( -25.20) >DroPer_CAF1 17984 116 + 1 ACCCCAAGUAGAAAAGAGGAAUACAAAGCGUGAUGUAUACGAGCGGGGAGUAAAUGGCUCAUUCAAUAUGCAUGAGCACAUUAAAUGAAAAACUCG--CAGCUCCAAUG--AUGGGGAGC .(((((............(.(((((........))))).)((((.(.((((.((((((((((.(.....).)))))).))))...(....))))).--).)))).....--.)))))... ( -32.70) >consensus ACCCCAACUAGAAUAGAGGAAAACAAAGCCCGACGCGCAUAAACGGGGAGUAAAUGGCUCAUACAAUAUGCAUGAGCACAUUAAAUGAACAGCACC__CAGCCCCAAGA_AAAGGGGAGC .((((......................((.....)).........(((.((.((((((((((.(.....).)))))).))))....(.....).......)))))........))))... (-15.48 = -15.12 + -0.36)


| Location | 17,333,554 – 17,333,672 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.87 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -18.06 |
| Energy contribution | -17.06 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.52 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17333554 118 - 27905053 GCUCCCCUUUUUCUUGGGGCUG--GGUGCUGUUCAUUUAAUGUGCUCAUGCAUAUUGUAUAAGCCAUUUGCUCCCCGUUUAUGCGCGUCGGGCUUUGAUGCCCUAUAAUCUAGUUGAGUU (((((((........)))((((--(((((.........(((((((....)))))))((((((((............))))))))))...((((......))))....))))))).)))). ( -36.50) >DroPse_CAF1 16978 116 - 1 GCUCCCCAU--CAUUGGAGCUG--CGAGUUUUUCAUUUAAUGUGCUCAUGCAUAUUGAAUGAGCCAUUUACUCCCCGCUCGUGUACAUCACGCUUUGUAUUCCUCUUUUCUAGUUGGGGU ...(((((.--(....((((.(--.((((..((((((((((((((....))))))))))))))......)))).).))))(.(((((........))))).)..........).))))). ( -37.60) >DroSec_CAF1 9522 118 - 1 GCUCCCCUUUUUCUUGGGGCUG--GGCGCUGUUCAUUUAAUGUGCUCAUGCAUAUUGUAUGAGCCAUUUGCUCCCCGUUUAUGCGCGUCGGGCUUUGAUGCCCUAUAAUCUAGUUGGGUU (((((((........)))((((--(((((.((((((.((((((((....)))))))).))))))(((..((.....))..))).)))))((((......)))).......)))).)))). ( -40.60) >DroAna_CAF1 13612 107 - 1 GCUCCCCUGUC-UUUGGGGCUGCGAGUGCUGCUCAUUUAAUGUUCUCAUGAAUUUUUUAUAAGCCAUUUACUCCCUGUUUAAGCCAG------------AAACUUCAGUCUAGUUGGGGU ...((((...(-(...((((((.(((((..(((.((..((.((((....)))).))..)).)))....))))).(((.......)))------------......))))))))..)))). ( -23.70) >DroPer_CAF1 17984 116 - 1 GCUCCCCAU--CAUUGGAGCUG--CGAGUUUUUCAUUUAAUGUGCUCAUGCAUAUUGAAUGAGCCAUUUACUCCCCGCUCGUAUACAUCACGCUUUGUAUUCCUCUUUUCUACUUGGGGU ...(((((.--.....((((.(--.((((..((((((((((((((....))))))))))))))......)))).).))))(.(((((........))))).)............))))). ( -36.70) >consensus GCUCCCCUUU_CAUUGGGGCUG__CGAGCUGUUCAUUUAAUGUGCUCAUGCAUAUUGUAUGAGCCAUUUACUCCCCGUUUAUGCACGUCACGCUUUGAAGCCCUAUAAUCUAGUUGGGGU (((((..........)))))...........((((...(((((((....)))))))((((((((............))))))))..............................)))).. (-18.06 = -17.06 + -1.00)


| Location | 17,333,594 – 17,333,699 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 88.39 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -23.39 |
| Energy contribution | -23.37 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17333594 105 - 27905053 ACCCCUCUC-CGUAAGCCUCGUAAAUAUGCUCCCCUUUUUCUUGGGGCUG--GGUGCUGUUCAUUUAAUGUGCUCAUGCAUAUUGUAUAAGCCAUUUGCUCCCCGUUU .........-....(((((((..((.............))..)))))))(--((.((.(((.((.((((((((....)))))))).)).))).....)).)))..... ( -24.22) >DroSec_CAF1 9562 103 - 1 CCCC--CUC-CGCAUGCCUCGUAAAUAUGCUCCCCUUUUUCUUGGGGCUG--GGCGCUGUUCAUUUAAUGUGCUCAUGCAUAUUGUAUGAGCCAUUUGCUCCCCGUUU ....--...-.(((((.(((((((((((((.((((........)))).((--(((((............))))))).))))))).)))))).))..)))......... ( -32.40) >DroSim_CAF1 15186 103 - 1 CCCC--CUC-CGCAUGCCUCGUAAAUAUGCUCCCCUUUUUCUUGGGGCUG--GGCGCUGUUCAUUUAAUGUGCUCAUGCAUAUUGUAUGAGCUAUUUGCUCCCCGUUU ....--...-.(((((.........))))).((((........))))..(--((.((.((((((.((((((((....)))))))).)))))).....)).)))..... ( -32.10) >DroEre_CAF1 14772 101 - 1 -CCC--CUC-CGUGAGCCUCGUAAAUAUGCUCCCCUUUUUCUUGGGGCUG---GUGCUGUUCAUUUAAUGUGCUCAUGCAUAUUGUAUGAGCCAUUUGCUCCCCGUUU -...--...-...((((...........))))...........(((((..---(((..((((((.((((((((....)))))))).)))))))))..)))))...... ( -28.80) >DroYak_CAF1 13110 102 - 1 -UCC--CUC-CGCAAGCCUCGUAAAUAUGCUCCCCUUUUCCUUGGGGCUG--GGUGCUGUUCAUUUAAUGUGCCCAUGCAUAUUGUAUGAGCCAUUUGCUCGCCGUUU -...--...-.(((((.(((((((((((((.((((........)))).((--((..(............)..)))).))))))).))))))...)))))......... ( -33.20) >DroAna_CAF1 13640 104 - 1 -CCC--CUCAGGUAAGCCCCAUAAAUACGCUCCCCUGUC-UUUGGGGCUGCGAGUGCUGCUCAUUUAAUGUUCUCAUGAAUUUUUUAUAAGCCAUUUACUCCCUGUUU -...--..((((..(((((((.....(((......))).-..)))))))..(((((..(((.((..((.((((....)))).))..)).)))....)))))))))... ( -26.10) >consensus _CCC__CUC_CGCAAGCCUCGUAAAUAUGCUCCCCUUUUUCUUGGGGCUG__GGUGCUGUUCAUUUAAUGUGCUCAUGCAUAUUGUAUGAGCCAUUUGCUCCCCGUUU ....................((((((.((((((((........)))).....))))..((((((.((((((((....)))))))).)))))).))))))......... (-23.39 = -23.37 + -0.02)



| Location | 17,333,699 – 17,333,815 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.85 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17333699 116 - 27905053 UGCCUUAAUGCGACUCUAAGCCAUAUAGUUAGCCAGGCAUUGCAACUGCCAUAAAAAACUUGGCAAUUAGCUUGUUUAACAAGAGCG-CUCUUUGUGGCUACAAAAUGCAGC-UACCU-- ........((((......(((((((..(((((.(((((........(((((.........)))))....))))).)))))((((...-.)))))))))))......))))..-.....-- ( -27.50) >DroSec_CAF1 9665 116 - 1 UGUCCUAAUACGACUCUAAGCCAUAUAGUUAGCCAGGCAUUGCAACUGCCAUAAAAAACUUGGCAAUUAGCUUGUUUAACAAGAGCG-CUCUUUGUGGCUACAAAAUGCAGC-UACCU-- .(((.......)))....(((((((..(((((.(((((........(((((.........)))))....))))).)))))((((...-.)))))))))))............-.....-- ( -25.40) >DroSim_CAF1 15289 116 - 1 UGUCCUAAUGCGACUCUAAGCCAUAUAGUUAGCCAGGCAUUGCAACUGCCAUAAAAAACUUGGCAAUUAGCUUGUUUAACAAGAGCG-CUCUUUGUGGCUACAAAAUGCAGC-CACCU-- .........(((.((((..(((.............)))...((((.(((((.........))))).)).))..........))))))-).....((((((.(.....).)))-)))..-- ( -29.12) >DroEre_CAF1 14873 102 - 1 UACCUUAAUGCGACUUUAAGCCAUAGAGUUAGCCAGU------------CAUAAAAAACUUGGCAAUUAGCUUGUUUAACAAGAGCU-CUCUUUGUGGCUACAAAAUGCAGCUCC----- ........((((......((((((((((..(((..((------------((.........))))......((((.....)))).)))-..))))))))))......)))).....----- ( -27.20) >DroYak_CAF1 13212 113 - 1 UACCUCAAUGGUACAUUGAGCCAUAUAGUUAGCCAGGCAUUGCGACUGGCAUAAAAAACUUGGCAAUUAGCUUGUUAAACAAGAGCG-CUCUUUGUGGCUACAAAAUGCAGC-CA----- .(((.....)))(((..((((....(((((.((((((...(((.....))).......)))))))))))((((.........)))))-)))..)))((((.(.....).)))-).----- ( -31.60) >DroPer_CAF1 18133 111 - 1 UAC----AACAUGUAUUUAUCCACUUGGUUAGUUAGAAACGGUUAAUGGC----AAAUGGUGCCAUUUAGCUUGUUCAACAAGAGUGGCUCUUUGUGGCUACAAAAUGCAGC-UAACCCC ...----...................((((((((.(((..((((((((((----(.....))))).))))))..))).......((((((......)))))).......)))-))))).. ( -29.90) >consensus UACCUUAAUGCGACUCUAAGCCAUAUAGUUAGCCAGGCAUUGCAACUGCCAUAAAAAACUUGGCAAUUAGCUUGUUUAACAAGAGCG_CUCUUUGUGGCUACAAAAUGCAGC_CACCU__ ..................(((((((.((...((((((.....................)))))).....((((.........))))....)).))))))).................... (-16.40 = -16.85 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:07 2006