
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,319,086 – 17,319,304 |
| Length | 218 |
| Max. P | 0.999149 |

| Location | 17,319,086 – 17,319,202 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -16.37 |
| Energy contribution | -17.17 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17319086 116 + 27905053 AAACUUGUUUAUGAUUUGCUUGAAGAAAUAAACACAUUUUAAUUAACAUUUAAAAUGCAUUUUAAGCAAAAAGCUUUUAAGAAA-CGGCGCCAUAACAAUAAUCAUAAC---AGCACAAA ....(((((.(((((((((((((((.........((((((((.......))))))))..)))))))))))..(((.((....))-.))).............)))))))---))...... ( -18.00) >DroSim_CAF1 606 116 + 1 AAACUUGUUAAUGAUUUGCUUGAAGAAAUAAACACAUUUUAAUUAACAUUUAAAAUGCGUUUUAAGCAAAAAGCUUUUAAGAAA-CGGCGCCAUAACAAUAAUCAUAAC---AGCACAAA ....((((((.(((((((((((((.......((.((((((((.......)))))))).))))))))))))..(((.((....))-.))).............)))))))---))...... ( -20.71) >DroEre_CAF1 537 120 + 1 AAACUUGUUAAUGAUCUGCUCGAAGAAAUAAACACAUUUUAAUUAACAUUUAAAAUGCGUUUUAAGCAAAAAGCUUUUAAGAAAUCGGCGCCAUAACAAUAAUCAUAACAAAAGCACAAA ....((((((.((((..((.(((......((((.((((((((.......)))))))).)))).((((.....))))........)))))(......)....))))))))))......... ( -18.70) >DroYak_CAF1 467 120 + 1 AAACUUGUUAAUGAUUUGCUUGAAGAAAUAAACACAUUUUAAUUAACAUUUAAAAUGCGUUUUAAGCAAAAAGCUUUUAAGAAAUCGGCGCCAUAACAAUAAUCAUAACAAAAGCACAAA ....((((((.(((((((((((((.......((.((((((((.......)))))))).))))))))))))..(((...........))).............)))))))))......... ( -20.71) >DroAna_CAF1 1290 119 + 1 AAACUUGUUAAUGAUUUGCUUUAAGAAAUAAACACAUUUUAAUUAACAUUUAAAAUGCGUUUUAAGCAAAAAGCUUUUAAGAAA-CGGCGCCAUAACAAUAAUCAUAAUAAAAGCACAAG ...(((((......(((((((........((((.((((((((.......)))))))).)))).)))))))..(((((((.....-.......................)))))))))))) ( -20.39) >consensus AAACUUGUUAAUGAUUUGCUUGAAGAAAUAAACACAUUUUAAUUAACAUUUAAAAUGCGUUUUAAGCAAAAAGCUUUUAAGAAA_CGGCGCCAUAACAAUAAUCAUAACAAAAGCACAAA ....(((((.(((.((((((((((.......((.((((((((.......)))))))).))))))))))))..(((...........)))..))))))))..................... (-16.37 = -17.17 + 0.80)

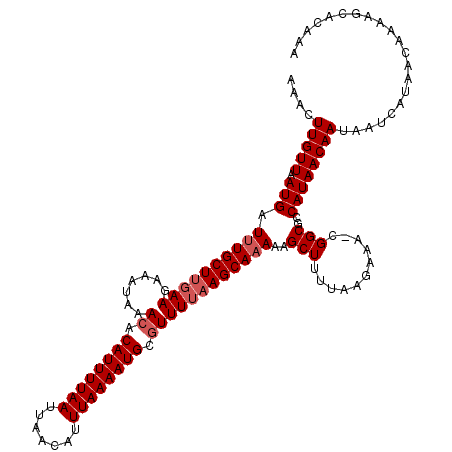
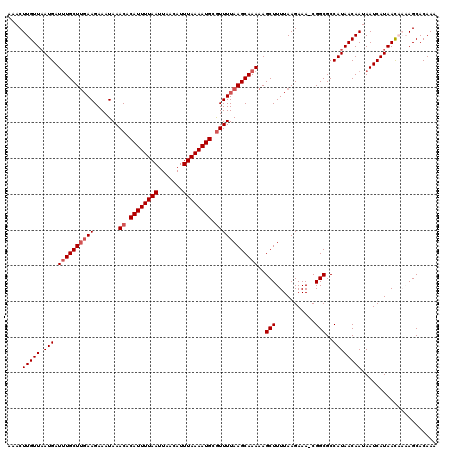
| Location | 17,319,086 – 17,319,202 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -22.58 |
| Energy contribution | -22.46 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17319086 116 - 27905053 UUUGUGCU---GUUAUGAUUAUUGUUAUGGCGCCG-UUUCUUAAAAGCUUUUUGCUUAAAAUGCAUUUUAAAUGUUAAUUAAAAUGUGUUUAUUUCUUCAAGCAAAUCAUAAACAAGUUU .....(((---.((((((((..((((.(((.....-........((((.....)))).(((((((((((((.......)))))))))))))......)))))))))))))))...))).. ( -21.70) >DroSim_CAF1 606 116 - 1 UUUGUGCU---GUUAUGAUUAUUGUUAUGGCGCCG-UUUCUUAAAAGCUUUUUGCUUAAAACGCAUUUUAAAUGUUAAUUAAAAUGUGUUUAUUUCUUCAAGCAAAUCAUUAACAAGUUU (((((((.---((((((((....)))))))))).(-(((.....((((.....)))).(((((((((((((.......)))))))))))))........)))).........)))))... ( -24.10) >DroEre_CAF1 537 120 - 1 UUUGUGCUUUUGUUAUGAUUAUUGUUAUGGCGCCGAUUUCUUAAAAGCUUUUUGCUUAAAACGCAUUUUAAAUGUUAAUUAAAAUGUGUUUAUUUCUUCGAGCAGAUCAUUAACAAGUUU ........((((((((((((...((....))(((((........((((.....)))).(((((((((((((.......)))))))))))))......))).)).))))).)))))))... ( -27.20) >DroYak_CAF1 467 120 - 1 UUUGUGCUUUUGUUAUGAUUAUUGUUAUGGCGCCGAUUUCUUAAAAGCUUUUUGCUUAAAACGCAUUUUAAAUGUUAAUUAAAAUGUGUUUAUUUCUUCAAGCAAAUCAUUAACAAGUUU (((((......((((((((....))))))))...((((((((..((((.....)))).(((((((((((((.......)))))))))))))........))).)))))....)))))... ( -26.50) >DroAna_CAF1 1290 119 - 1 CUUGUGCUUUUAUUAUGAUUAUUGUUAUGGCGCCG-UUUCUUAAAAGCUUUUUGCUUAAAACGCAUUUUAAAUGUUAAUUAAAAUGUGUUUAUUUCUUAAAGCAAAUCAUUAACAAGUUU (((((((((((((((((((....))))))).....-.....)))))))..(((((((.(((((((((((((.......)))))))))))))........)))))))......)))))... ( -24.30) >consensus UUUGUGCUUUUGUUAUGAUUAUUGUUAUGGCGCCG_UUUCUUAAAAGCUUUUUGCUUAAAACGCAUUUUAAAUGUUAAUUAAAAUGUGUUUAUUUCUUCAAGCAAAUCAUUAACAAGUUU (((((......((((((((....))))))))((...........((((.....)))).(((((((((((((.......)))))))))))))..........)).........)))))... (-22.58 = -22.46 + -0.12)

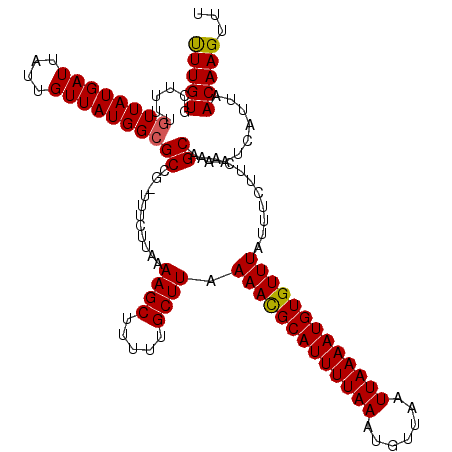

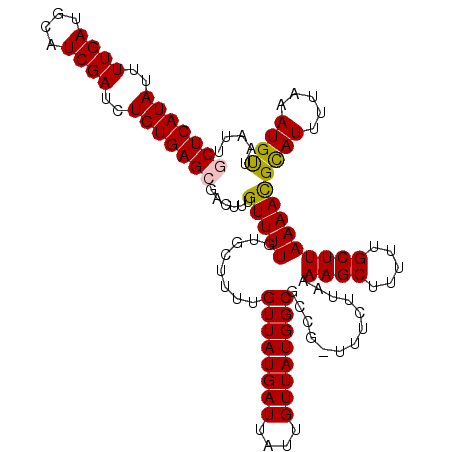
| Location | 17,319,126 – 17,319,240 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.13 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
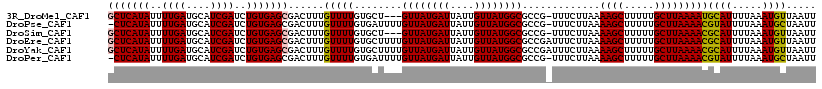
>3R_DroMel_CAF1 17319126 114 - 27905053 GCUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGUUUUGUGCU---GUUAUGAUUAUUGUUAUGGCGCCG-UUUCUUAAAAGCUUUUUGCUUAAAAUGCAUUUUAAAUGUUAAUU (((((((..((((....))))..)))))))(((((((....((((.---((((((((....))))))))....-........((((.....)))).....))))..)))).))).... ( -27.10) >DroPse_CAF1 2951 116 - 1 -CUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGUUUUGUGAUUUUGUUAUGAUUAUUGUUAUGGCGCCG-UUUCUUAAAAGCUUUUUGCUUAAAACGUAUUUUAAAUGCUAAUU -((((((..((((....))))..)))))).......(((((........((((((((....))))))))....-........((((.....)))))))))((((.....))))..... ( -20.00) >DroSim_CAF1 646 114 - 1 GCUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGUUUUGUGCU---GUUAUGAUUAUUGUUAUGGCGCCG-UUUCUUAAAAGCUUUUUGCUUAAAACGCAUUUUAAAUGUUAAUU (((((((..((((....))))..)))))))(((((((....((((.---((((((((....))))))))...(-(((.....((((.....)))).))))))))..)))).))).... ( -27.50) >DroEre_CAF1 577 118 - 1 GCUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGUUUUGUGCUUUUGUUAUGAUUAUUGUUAUGGCGCCGAUUUCUUAAAAGCUUUUUGCUUAAAACGCAUUUUAAAUGUUAAUU (((((((..((((....))))..)))))))(((((((...((((.((((((((((((....)))))))).............((((.....))))))))))))...)))).))).... ( -27.50) >DroYak_CAF1 507 118 - 1 GCUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGUUUUGUGCUUUUGUUAUGAUUAUUGUUAUGGCGCCGAUUUCUUAAAAGCUUUUUGCUUAAAACGCAUUUUAAAUGUUAAUU (((((((..((((....))))..)))))))(((((((...((((.((((((((((((....)))))))).............((((.....))))))))))))...)))).))).... ( -27.50) >DroPer_CAF1 2980 116 - 1 -CUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGUUUUGUGAUUUUGUUAUGAUUAUUGUUAUGGCGCCG-UUUCUUAAAAGCUUUUUGCUUAAAACGUAUUUUAAAUGCUAAUU -((((((..((((....))))..)))))).......(((((........((((((((....))))))))....-........((((.....)))))))))((((.....))))..... ( -20.00) >consensus GCUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGUUUUGUGCUUUUGUUAUGAUUAUUGUUAUGGCGCCG_UUUCUUAAAAGCUUUUUGCUUAAAACGCAUUUUAAAUGUUAAUU (((((((..((((....))))..)))))))......(((((........((((((((....)))))))).............((((.....)))))))))((((.....))))..... (-22.38 = -22.13 + -0.25)
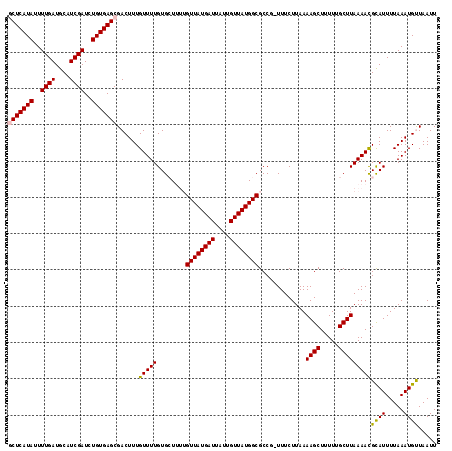
| Location | 17,319,202 – 17,319,304 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -22.27 |
| Energy contribution | -23.02 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17319202 102 + 27905053 ACAAAGUCGCUCACAGAUCGAUGCAUCAAAAUAUGAGCGGCGAGGGGCACGGAACGUGGCUUUUCGUUGAUUCCAGGGGAGUCUCGACCCCAAGUUCCCCGA .....((((((((......((....))......))))))))..((((.((((((..((((.....)))).)))).((((........))))..)).)))).. ( -33.50) >DroSim_CAF1 722 96 + 1 ACAAAGUCGCUCACAGAUCGAUGCAUCAAAAUAUGAGCGGCGAGGGGCACGGCACGAAAAGCC------CCUCCAGGGGAGUCUCGACCCCCAGUUCUGCCU .....((((((((......((....))......))))))))(((((((............)))------))))..((((.((....)))))).......... ( -34.80) >DroEre_CAF1 657 94 + 1 ACAAAGUCGCUCACAGAUCGAUGCAUCAAAAUAUGAGCGGUGG--GGCACGGAACGAAAAGCC------ACUCCAGGGGAAUCUCGACCCCGAGUCCCUCAA .......((((((......((....))......))))))(.((--(((..(((..(......)------..))).((((........))))..))))).).. ( -26.30) >DroYak_CAF1 587 96 + 1 ACAAAGUCGCUCACAGAUCGAUGCAUCAAAAUAUGAGCGGUGGGGGGCAAAGAACGAAAUGCC------ACUCCAGGGGGAUCUCGACCCCGAGCUCCUCAA ......(((((((......((....))......)))))))(((((((((..........))))------.))))).(((((.((((....)))).))))).. ( -32.20) >consensus ACAAAGUCGCUCACAGAUCGAUGCAUCAAAAUAUGAGCGGCGAGGGGCACGGAACGAAAAGCC______ACUCCAGGGGAAUCUCGACCCCGAGUUCCUCAA .....((((((((......((....))......))))))))(((((((..(((..................))).((((........))))..))))))).. (-22.27 = -23.02 + 0.75)


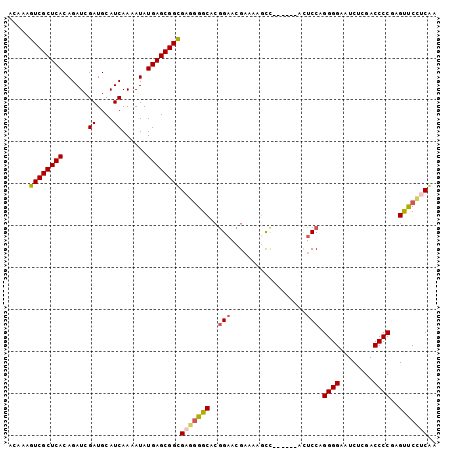
| Location | 17,319,202 – 17,319,304 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -35.39 |
| Consensus MFE | -24.46 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17319202 102 - 27905053 UCGGGGAACUUGGGGUCGAGACUCCCCUGGAAUCAACGAAAAGCCACGUUCCGUGCCCCUCGCCGCUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGU ..((((.((..((((........)))).(((((..............))))))).))))....((((((((..((((....))))..))))))))....... ( -33.94) >DroSim_CAF1 722 96 - 1 AGGCAGAACUGGGGGUCGAGACUCCCCUGGAGG------GGCUUUUCGUGCCGUGCCCCUCGCCGCUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGU .(((......((((((....))))))...((((------(((............))))))))))(((((((..((((....))))..)))))))........ ( -42.40) >DroEre_CAF1 657 94 - 1 UUGAGGGACUCGGGGUCGAGAUUCCCCUGGAGU------GGCUUUUCGUUCCGUGCC--CCACCGCUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGU ....(((.(.(((((.(((((..((.(....).------))..)))))))))).).)--))..((((((((..((((....))))..))))))))....... ( -34.20) >DroYak_CAF1 587 96 - 1 UUGAGGAGCUCGGGGUCGAGAUCCCCCUGGAGU------GGCAUUUCGUUCUUUGCCCCCCACCGCUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGU .((.((.((((.(((..(....)..))).))))------((((..........)))))).)).((((((((..((((....))))..))))))))....... ( -31.00) >consensus UUGAGGAACUCGGGGUCGAGACUCCCCUGGAGU______GGCUUUUCGUUCCGUGCCCCCCACCGCUCAUAUUUUGAUGCAUCGAUCUGUGAGCGACUUUGU ..(((((((..((((........)))).(((((.......)))))..))))))).........((((((((..((((....))))..))))))))....... (-24.46 = -25.40 + 0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:55 2006