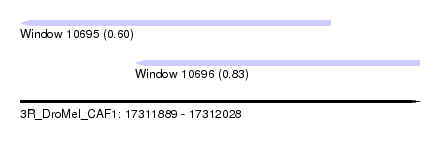
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,311,889 – 17,312,028 |
| Length | 139 |
| Max. P | 0.832789 |

| Location | 17,311,889 – 17,311,997 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -43.42 |
| Consensus MFE | -28.13 |
| Energy contribution | -27.83 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
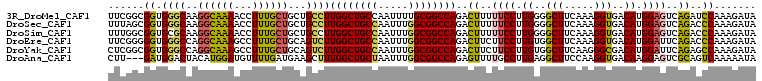
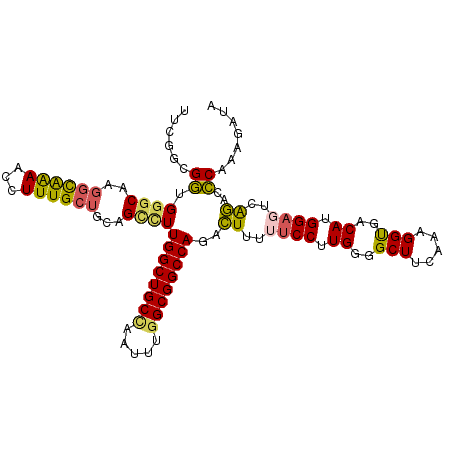
>3R_DroMel_CAF1 17311889 108 - 27905053 UUCGGCGGUGGGCAAGGCAAAACCUUUGCUGCUGCCUUGGCUGCCAAUUUUGCGGCCAGACUUUUUCCUUGGGGCUUCAAAGGUGACAUGGAGUCAGAUCCAAAGAUA ...(((((..(.(((((......))))))..)))))((((((((.......))))))))......((.(((((((((((..(....).)))))))....)))).)).. ( -40.20) >DroSec_CAF1 44485 108 - 1 UUUAGCGGUGGGCAAGGCAAAACCUUUGCUGCUGCCUUGGCUGCCAAUUUGGCGGCCAGACUUUUUCCUUGGGGCUUCAAAGGUGACAUGGAGUCAGACCCAAAGAUA ....((((..(.(((((......))))))..)))).(((((((((.....)))))))))......((.(((((((((((..(....).))))))....))))).)).. ( -42.30) >DroSim_CAF1 46202 108 - 1 UUUGGCGGUGCGCAAGGCAAAACCUUUGCUGCUGCCUUGGCUGCCAAUUUGGCGGCCAGACUUUUUCCUUGGGGCUUCAAAGGUGACAUGGAGUCAGACCCAAAGAUA ...(((((((.((((((......)))))))))))))(((((((((.....)))))))))......((.(((((((((((..(....).))))))....))))).)).. ( -45.70) >DroEre_CAF1 46393 108 - 1 UUCGGGGGUGGGCCAGGCAAAGCCUUUGCUGCAGUCUUGGCUGCCAAUUUGGCGGCCAGACUUCUUCCUUGUGGCUUCAAAGGUGACAUGGAUUCAGACCCAAAGAUA ...(((..(((((((......(((((((..(((((((.(((((((.....)))))))))))).....(....)))..)))))))....))).))))..)))....... ( -44.80) >DroYak_CAF1 49272 108 - 1 CUCGGCGGUGGGCCAGGCAAAGCCUUUGCUGCAGUCUUGGCUGCCAAUUUGGCGGCCAGACUUCUUCCUUGUGGCUUCAAGGGCGACAUGGAUUCAGAGCCAAAGAUA ...(((..(((((((......(((((((..(((((((.(((((((.....)))))))))))).....(....)))..)))))))....))).))))..)))....... ( -45.30) >DroAna_CAF1 44689 105 - 1 CUU---GAUGGACUACAUGGAUGUUUUGAUGAAGCUUUGGCUGCUAAUUUGGCGGCCAGAGUUUUGCCUUGAGGCUUCCAAGGUGACAAGGAGUCGCAGUCAAAAAUA ...---....((((.(.((((.((((..(((((((((((((((((.....))))))))))))))))..)..)))).)))).)(((((.....)))))))))....... ( -42.20) >consensus UUCGGCGGUGGGCAAGGCAAAACCUUUGCUGCAGCCUUGGCUGCCAAUUUGGCGGCCAGACUUUUUCCUUGGGGCUUCAAAGGUGACAUGGAGUCAGACCCAAAGAUA ......((.((((..((((((...))))))...))))((((((((.....))))))))..((..((((.((..(((.....)))..)).))))..))..))....... (-28.13 = -27.83 + -0.30)

| Location | 17,311,929 – 17,312,028 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -25.65 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
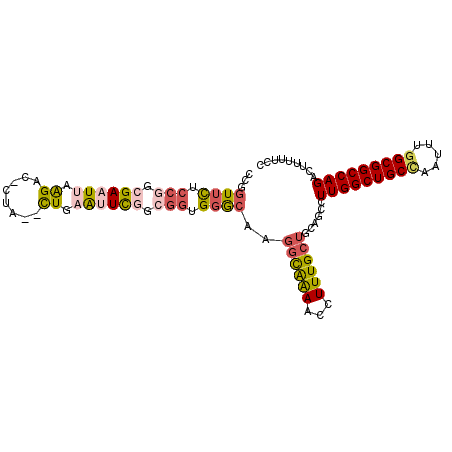
>3R_DroMel_CAF1 17311929 99 - 27905053 CCGGUUCUCCGGCGAAUUAAGAC-CUA--CUGAAUUCGGCGGUGGGCAAGGCAAAACCUUUGCUGCUGCCUUGGCUGCCAAUUUUGCGGCCAGACUUUUUCC ((((....)))).(((((.((..-...--)).)))))(((((..(.(((((......))))))..)))))((((((((.......))))))))......... ( -36.80) >DroSec_CAF1 44525 99 - 1 CCGGUUCUCCGGCGAAUUAAGAC-CUG--CUGAAUUUAGCGGUGGGCAAGGCAAAACCUUUGCUGCUGCCUUGGCUGCCAAUUUGGCGGCCAGACUUUUUCC ((((....)))).(((..(((((-(.(--(((....)))))))(((((.(((((.....)))))..)))))((((((((.....))))))))..))).))). ( -40.00) >DroSim_CAF1 46242 99 - 1 CCGGUUCUCCGGCGAAUUAAGAC-CUG--CUGAAUUUGGCGGUGCGCAAGGCAAAACCUUUGCUGCUGCCUUGGCUGCCAAUUUGGCGGCCAGACUUUUUCC ((((....)))).(((..(((..-...--........(((((((.((((((......)))))))))))))(((((((((.....))))))))).))).))). ( -39.50) >DroEre_CAF1 46433 99 - 1 UCGGUUCUCCGGCGAAUUAAGAC-CUA--UUGAAUUCGGGGGUGGGCCAGGCAAAGCCUUUGCUGCAGUCUUGGCUGCCAAUUUGGCGGCCAGACUUCUUCC ..(((((.((..((((((((...-...--))).)))))..)).))))).(((((.....)))))(.(((((.(((((((.....)))))))))))).).... ( -38.50) >DroYak_CAF1 49312 99 - 1 CCGGUUCUCCGGCGAAUUAAGAC-CUA--CUGCACUCGGCGGUGGGCCAGGCAAAGCCUUUGCUGCAGUCUUGGCUGCCAAUUUGGCGGCCAGACUUCUUCC .(((....)))(((((....(.(-(((--((((.....)))))))).).(((...)))))))).(.(((((.(((((((.....)))))))))))).).... ( -42.50) >DroAna_CAF1 44729 99 - 1 CCGAUUUUCGGGAGAUUGUGGGCACUUCUUUGAGCUU---GAUGGACUACAUGGAUGUUUUGAUGAAGCUUUGGCUGCUAAUUUGGCGGCCAGAGUUUUGCC (((.....))).........(((...((...((((((---.(((.....))).)).)))).)).(((((((((((((((.....)))))))))))))))))) ( -34.80) >consensus CCGGUUCUCCGGCGAAUUAAGAC_CUA__CUGAAUUCGGCGGUGGGCAAGGCAAAACCUUUGCUGCAGCCUUGGCUGCCAAUUUGGCGGCCAGACUUUUUCC ...((((.(((.((((((.((........)).)))))).))).))))..((((((...))))))......(((((((((.....)))))))))......... (-25.65 = -26.60 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:47 2006