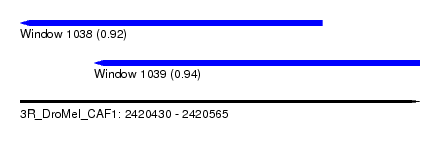
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,420,430 – 2,420,565 |
| Length | 135 |
| Max. P | 0.940312 |

| Location | 2,420,430 – 2,420,532 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.38 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -22.33 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
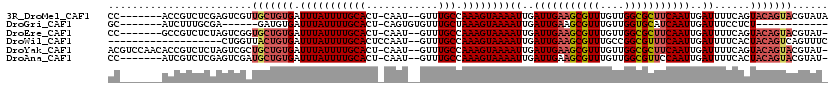
>3R_DroMel_CAF1 2420430 102 - 27905053 GCACU-CAAUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAUACGAAUACGUAUGUUGCAGCACCCCU--------------- ((...-..............(.(((((..(((((((((((....)))))))))))..)))))).(((..((((((((....))))))))..))).))......--------------- ( -32.90) >DroSim_CAF1 125606 102 - 1 GCACU-CAAUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAUACGAAUACGUAUGUUGCAGCACCCCU--------------- ((...-..............(.(((((..(((((((((((....)))))))))))..)))))).(((..((((((((....))))))))..))).))......--------------- ( -32.90) >DroEre_CAF1 128362 94 - 1 GCACU-CAAUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAU------ACGUAUGUUGCAGCACCC----------------- ((...-..............(.(((((..(((((((((((....)))))))))))..)))))).(((..(((((...------.)))))..))).))....----------------- ( -29.70) >DroWil_CAF1 196312 97 - 1 GCACUCCAAUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGCCGGCGUUUCAAUUGAUUUUCACUACAGUCAGUUUCC---UUCACAUCCCAUAGAA---CU--------------- (((..(....)..)))...((((((((..((((((((((......))))))))))..))))).))).............---................---..--------------- ( -20.30) >DroYak_CAF1 122632 111 - 1 GCACU-CAAUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAU------ACGUAUGUUGCAGCACCUCUCCACCCCUCCACCCC ((...-..............(.(((((..(((((((((((....)))))))))))..)))))).(((..(((((...------.)))))..))).))..................... ( -29.70) >DroAna_CAF1 125773 96 - 1 GCACU-CAAUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGUUCCAAUUGAUUUUCACUACAGUACGUAU------CCGUGUGUUGCACCGCCACA--------------- ((((.-..(((..(((...((((((((..((((.((((((....)))))).))))..))))).)))...)))..)))------..))))((.((...)).)).--------------- ( -21.60) >consensus GCACU_CAAUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAU______ACGUAUGUUGCAGCACCCCU_______________ (((.................(.(((((..(((((((((((....)))))))))))..))))))......((((((........))))))..)))........................ (-22.33 = -23.00 + 0.67)

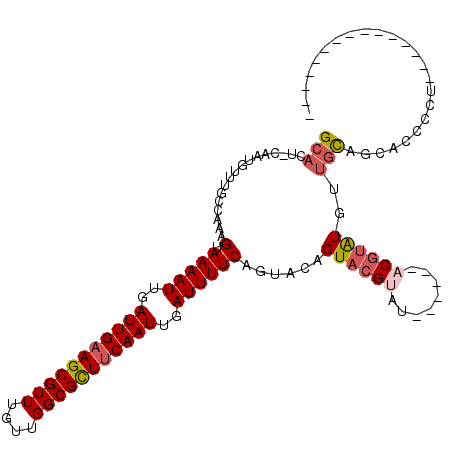
| Location | 2,420,455 – 2,420,565 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -23.21 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2420455 110 - 27905053 CC-------ACCGUCUCGAGUCGUUGCUGUGAUUUAUUUUGCACU-CAAU--GUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAUA ..-------............(((.((((((.(((((((((((..-....--...)).)))))))))((..(((((((((((....)))))))))))..))......))))))))).... ( -30.80) >DroGri_CAF1 201438 94 - 1 GC-------AUCUUUGCGA------GAUGUGAUUUAUUUUGCACU-CAGUGUGUUUGCUAAAGUAAAAUUGAUUGAAGCGUUUGUUGGUGCAUCAAUUGAUUUCCUCU------------ ((-------(((((...))------)))))..(((((((((((..-(.....)..))).))))))))((..(((((.(((........))).)))))..)).......------------ ( -21.10) >DroEre_CAF1 128380 109 - 1 CC-------GCCGUCUCUAGUCGGUGCUGUGAUUUAUUUUGCACU-CAAU--GUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAU- ..-------((((.(....).))))((((((.(((((((((((..-....--...)).)))))))))((..(((((((((((....)))))))))))..))......))))))......- ( -34.20) >DroWil_CAF1 196331 99 - 1 -------------------CUGGUUACUGUGAUUUAUUUUGCACUCCAAU--GUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGCCGGCGUUUCAAUUGAUUUUCACUACAGUCAGUUUC -------------------(((..(...(((((((((((((((..(....--)..)).)))))))))((..((((((((((......))))))))))..))..))))...)..))).... ( -27.50) >DroYak_CAF1 122667 116 - 1 ACGUCCAACACCGUCUCUAGUCGCUGCUGUGAUUUAUUUUGCACU-CAAU--GUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAU- .....................((.(((((((.(((((((((((..-....--...)).)))))))))((..(((((((((((....)))))))))))..))......)))))))))...- ( -30.80) >DroAna_CAF1 125793 109 - 1 CC-------AUCGUCUCGAGUCGAUGCUGUGAUUUAUUUUGCACU-CAAU--GUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGUUCCAAUUGAUUUUCACUACAGUACGUAU- .(-------((((.(....).)))))(((((.(((((((((((..-....--...)).)))))))))((..((((.((((((....)))))).))))..))......))))).......- ( -25.20) >consensus CC_______ACCGUCUCGAGUCGUUGCUGUGAUUUAUUUUGCACU_CAAU__GUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCACUACAGUACGUAU_ ........................(((((((.(((((((((((............))).))))))))((..(((((((((((....)))))))))))..))......)))))))...... (-23.21 = -24.12 + 0.91)
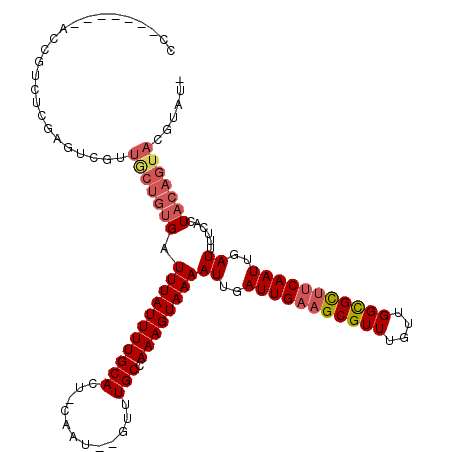
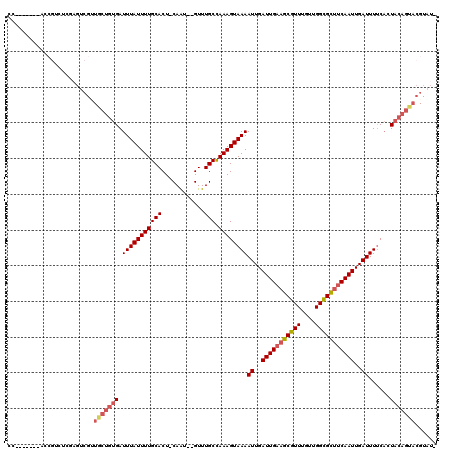
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:31 2006