| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,420,284 – 2,420,398 |
| Length | 114 |
| Max. P | 0.610729 |

| Location | 2,420,284 – 2,420,398 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.14 |
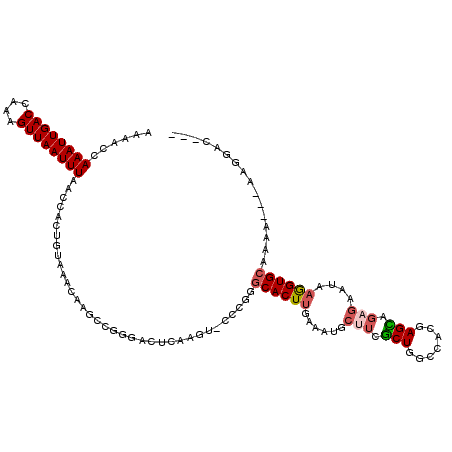
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -13.41 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2420284 114 + 27905053 AAAACCAAAUUGACCAAAGUUAAUUUAACCACUGUAAACAAGCCGGGACUCAAGUGCCCGGGCACUUGAAAUGCUUCGCUGGCCACGAGCAGAGAAUAAGGUGCAAAA---AUGGAC--- ......((((((((....))))))))..(((...........(((((((....)).)))))((((((......((..(((.(...).)))..))....))))))....---.)))..--- ( -31.00) >DroSim_CAF1 125459 114 + 1 AAAACCAAAUUGACCAAAGUUAAUUUAACCACUGUAAACAAGCCGGGACUCAAGUGCCCGGGCACUUGAAAUGCUUCGCUGGCCACGAGCAGAGAAUAAGGUGCAAAA---AAGGAC--- ...(((((((((((....)))))))).....((((......(((((((.((((((((....))))))))......)).))))).....)))).......)))......---......--- ( -30.50) >DroEre_CAF1 128217 114 + 1 AAAACCAAAUUGACCAAAGUUAAUUUAACCACUGUAAACAAGCCGGGACUCAAGUACCUGGGCACUUGAAAUGCUUCGCUGGCCACGAGCAGAGAAUAAGGUGCAAAA---AAGGAC--- ......((((((((....))))))))..((............(((((((....)).)))))((((((......((..(((.(...).)))..))....))))))....---..))..--- ( -28.30) >DroWil_CAF1 196168 92 + 1 AAAACCAAAUUGACCAAAGUUAAUUUAACCACUGUAAACAAGAC---------------AAGCACUUCAAAUGCAUCUCUGGGCAUGAGA-GA-AAGAAAGUGGAAGA--------C--- ......((((((((....))))))))..(((((...........---------------..(((.......))).(((((.......)))-))-.....)))))....--------.--- ( -17.40) >DroYak_CAF1 122477 117 + 1 AAAACCAAAUUGACCAAAGUUAAUUUAACCACUGUAAACAAGCAGGGACUCAAGUACCCGGGCACUUGAAAUGCUUCGCUGCCCACCAGCAGAGAAUAAGGUGCAAAAAAAAAAGAC--- ....((((((((((....))))))))..((.((((......))))))............))((((((......((..((((.....))))..))....)))))).............--- ( -28.20) >DroAna_CAF1 125610 108 + 1 AAAACCAAAUUGACCAAAGUUAAUUUAACCACUGUAAACACUCCCUGACUCGAA------GGCACUCGAAACGCUUCGCUGGCCACGAGU---GAGGAAAGUGCAAAA---AAGAAGUCC ......((((((((....)))))))).....((.....((((.(((.(((((..------(((...((((....))))...))).)))))---.)))..)))).....---.))...... ( -27.50) >consensus AAAACCAAAUUGACCAAAGUUAAUUUAACCACUGUAAACAAGCCGGGACUCAAGU_CCCGGGCACUUGAAAUGCUUCGCUGGCCACGAGCAGAGAAUAAGGUGCAAAA___AAGGAC___ ......((((((((....))))))))...................................((((((......(((.(((.......))).)))....))))))................ (-13.41 = -13.80 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:29 2006