| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,267,472 – 17,267,570 |
| Length | 98 |
| Max. P | 0.896499 |

| Location | 17,267,472 – 17,267,570 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -37.47 |
| Consensus MFE | -23.45 |
| Energy contribution | -23.83 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17267472 98 + 27905053 CCCUUGCCCUCUGCGUCGUGGCCAUGGAAGAUGUUAAGGAUGUCCUGGAUCUUCCUUUGGCC---GUCGAAUGCCUGGUGAAAAUGAAUUGGGCGAAA--AGA ...((((((...((((((((((((.(((((((....(((....)))..)))))))..)))))---).)).)))).(.((....)).)...))))))..--... ( -38.20) >DroSec_CAF1 164 98 + 1 CCCUUGCCCUCUGCGUCGUGGCCACGGAAGAUGUUAAGGAUGUCCUGGAUCUUCCUUUGGCC---GUCGAAUGUCUACAGGAAAUGCGUUGGGCGAAA--AUA ...((((((..(((((((((((((.(((((((....(((....)))..)))))))..)))))---).)))...((.....))..))))..))))))..--... ( -38.60) >DroSim_CAF1 164 98 + 1 CCCUUGCCCUCUGCGUCGUGGGCACGGAAGAUGUUAAGGAUGUCCUGGAUCUUCCUUUGGCC---GUCGAAUGCCUUCAGGAAAUGCGUUGGGCGAAA--AUA ...((((((..((((((((((.((.(((((((....(((....)))..)))))))..)).))---).)))...((....))...))))..))))))..--... ( -35.60) >DroEre_CAF1 1516 98 + 1 CCCUUGCCCUCUGCGUUGUGGCCUCGGAAGCUGUUAAGGAUGUCCUGCAUCUUCCUUUGGCC---GGCGAAUGUCUGGGGGAAAUGGGUUGGGCGAAA--AGA (((...(((((.(((((((((((..(((((.(((..(((....)))))).)))))...))))---.)).)))))..)))))....)))..........--... ( -37.70) >DroYak_CAF1 209 97 + 1 CCCUUGCCCUCUGCGUUGUGGCCUCGGAAGCUGUUCAGGAUGUCCUGCAUCUUCCU-UGGCC---GUCGAAUGCCUGGGGGAAAUGCGUUGGGCGAAA--AGA ...((((((..((((((((((((..(((((.((..((((....)))))).))))).-.))))---).).....((....)).))))))..))))))..--... ( -39.60) >DroPer_CAF1 34900 91 + 1 CCUUUGCCCUCUUUAU--UGGCCUCGGAAGAACUUCAGGAUGUCCUUGUGCUUCCCGGGGCCUAAGCCCAGUGC----------UGGGUCGGGGAACUCUUCU (((..((((.......--.(((((((((((.((...(((....))).)).))).))))))))..(((.....))----------)))))..)))......... ( -35.10) >consensus CCCUUGCCCUCUGCGUCGUGGCCACGGAAGAUGUUAAGGAUGUCCUGGAUCUUCCUUUGGCC___GUCGAAUGCCUGGAGGAAAUGCGUUGGGCGAAA__AGA ...((((((..(((((...((((..(((((......(((....)))....)))))...))))...........((....))..)))))..))))))....... (-23.45 = -23.83 + 0.38)

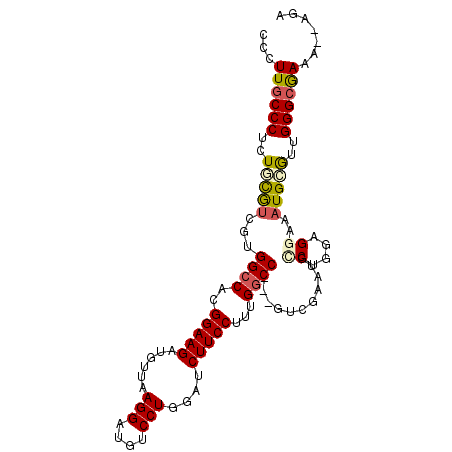
| Location | 17,267,472 – 17,267,570 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17267472 98 - 27905053 UCU--UUUCGCCCAAUUCAUUUUCACCAGGCAUUCGAC---GGCCAAAGGAAGAUCCAGGACAUCCUUAACAUCUUCCAUGGCCACGACGCAGAGGGCAAGGG .((--(((.((((..........(....)((..(((..---(((((..(((((((..(((....)))....))))))).))))).))).))...))))))))) ( -30.40) >DroSec_CAF1 164 98 - 1 UAU--UUUCGCCCAACGCAUUUCCUGUAGACAUUCGAC---GGCCAAAGGAAGAUCCAGGACAUCCUUAACAUCUUCCGUGGCCACGACGCAGAGGGCAAGGG ...--....((((...((......((....)).(((..---(((((..(((((((..(((....)))....))))))).))))).))).))...))))..... ( -31.50) >DroSim_CAF1 164 98 - 1 UAU--UUUCGCCCAACGCAUUUCCUGAAGGCAUUCGAC---GGCCAAAGGAAGAUCCAGGACAUCCUUAACAUCUUCCGUGCCCACGACGCAGAGGGCAAGGG ...--.....(((...............(((.......---.)))...(((((((..(((....)))....))))))).(((((.(......).))))).))) ( -28.70) >DroEre_CAF1 1516 98 - 1 UCU--UUUCGCCCAACCCAUUUCCCCCAGACAUUCGCC---GGCCAAAGGAAGAUGCAGGACAUCCUUAACAGCUUCCGAGGCCACAACGCAGAGGGCAAGGG .((--(((.((((...............(.....)...---((((...(((((.((.(((....)))...)).)))))..))))..........))))))))) ( -27.20) >DroYak_CAF1 209 97 - 1 UCU--UUUCGCCCAACGCAUUUCCCCCAGGCAUUCGAC---GGCCA-AGGAAGAUGCAGGACAUCCUGAACAGCUUCCGAGGCCACAACGCAGAGGGCAAGGG .((--(((.((((...((....((....))........---((((.-.(((((...((((....)))).....)))))..)))).....))...))))))))) ( -33.80) >DroPer_CAF1 34900 91 - 1 AGAAGAGUUCCCCGACCCA----------GCACUGGGCUUAGGCCCCGGGAAGCACAAGGACAUCCUGAAGUUCUUCCGAGGCCA--AUAAAGAGGGCAAAGG ..........((((.((((----------....)))))...((((.((((((((...(((....)))...))).))))).)))).--.......)))...... ( -30.60) >consensus UCU__UUUCGCCCAACGCAUUUCCCCCAGGCAUUCGAC___GGCCAAAGGAAGAUCCAGGACAUCCUUAACAUCUUCCGAGGCCACGACGCAGAGGGCAAGGG .........((((............................((((...(((((....(((....)))......)))))..)))).(......).))))..... (-21.27 = -21.77 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:29 2006