
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,262,951 – 17,263,143 |
| Length | 192 |
| Max. P | 0.998169 |

| Location | 17,262,951 – 17,263,063 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -39.57 |
| Consensus MFE | -33.92 |
| Energy contribution | -33.89 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17262951 112 + 27905053 CAGCGUUUAUCCACAAGAUACACACAAAAGUGCCGCGCCCUGCGGCACGGUUGAGAA-------UGGACCUCAACCGAAGUUUGGGGCAGUGGCAAUUUGUAUAUCUUUGUGAUAAAUC ....(((((((.((((((((...(((((..((((((((((((.(((.((((((((..-------.....))))))))..))))))))).)))))).))))).)))).))))))))))). ( -44.90) >DroSec_CAF1 59539 112 + 1 CAGCGUUUAUCCACAAGAUACACACAAAAGUGCCGCGCCCUGCGGCACGGUACAGAA-------UGGACCUCAACCGAAGUUCGGGGCAGUGGCAAUUUGUAUAUCUUUGUGAUAAAUC ....(((((((.((((((((...(((((..((((((((((((.(((.((((..((..-------.....))..))))..))))))))).)))))).))))).)))).))))))))))). ( -38.80) >DroSim_CAF1 62310 112 + 1 CAGCGUUUAUCCACAAGAUACACACAAAAGUGCCGCGCCCUGCGGCACGGUACAGAA-------UGGGCCUCAACCGAAGUUCGGGGCAGUGGCAAUUUGUAUAUCUUUGUGAUAAAUC ....(((((((.((((((((...(((((..((((((((((((.(((.((((..((..-------.....))..))))..))))))))).)))))).))))).)))).))))))))))). ( -38.80) >DroEre_CAF1 61310 112 + 1 CAGCGUUUAUCCACAAGAUACACACAAAAGUGCCGCGCCCUGCGGCACGGUACAGAA-------UGGACCUCAACCGAAGAUUGGGGCAGUGGCAAUUUGUAUAUCUUUGUGAUAAAUC ....(((((((.((((((((...(((((..(((((((((((.(((...(((.(....-------.).)))....)))......))))).)))))).))))).)))).))))))))))). ( -36.10) >DroYak_CAF1 82366 112 + 1 CAGCGUUUAUCCACAAGAUACACACAAAAGUGCCGCGCCCUGCGGCACGGUACAGAA-------UGGAGCUCAACCGAAGUUUGGGGCAGUGGCAAUUUGUAUAUCUUUGUGAUAAAUC ....(((((((.((((((((...(((((..((((((((((((.(((.((((..((..-------.....))..))))..))))))))).)))))).))))).)))).))))))))))). ( -35.90) >DroAna_CAF1 49675 117 + 1 CAGCGUUUAUCCAUAAGAUACACA-AAAAGUGCCGCGCCCUGCGGCAAGAUACAGAAUACAGCAUGGACCUCAGCCGGGGC-UGGGGCAGUGGCAAUUUGUAUAUCUUUGUGAUAAAUC ....(((((((((.((((((.(((-((...(((((((((....)))....................(.(((((((....))-)))))).)))))).))))).)))))))).))))))). ( -42.90) >consensus CAGCGUUUAUCCACAAGAUACACACAAAAGUGCCGCGCCCUGCGGCACGGUACAGAA_______UGGACCUCAACCGAAGUUUGGGGCAGUGGCAAUUUGUAUAUCUUUGUGAUAAAUC ....(((((((((.((((((.(((......((((((((((((.....((((..((..............))..)))).....)))))).))))))...))).)))))))).))))))). (-33.92 = -33.89 + -0.03)



| Location | 17,262,951 – 17,263,063 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -39.89 |
| Consensus MFE | -35.22 |
| Energy contribution | -35.13 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17262951 112 - 27905053 GAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAACUUCGGUUGAGGUCCA-------UUCUCAACCGUGCCGCAGGGCGCGGCACUUUUGUGUGUAUCUUGUGGAUAAACGCUG ..((((((...((((((((((...((((.(.((((.......(((((((((....-------.))))))))).......))))).))))......))))))))))...))))))..... ( -44.74) >DroSec_CAF1 59539 112 - 1 GAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCGAACUUCGGUUGAGGUCCA-------UUCUGUACCGUGCCGCAGGGCGCGGCACUUUUGUGUGUAUCUUGUGGAUAAACGCUG ..((((((...((((((((((...((((.(.((((.(.....((((..(((....-------.)))..)))).....).))))).))))......))))))))))...))))))..... ( -38.10) >DroSim_CAF1 62310 112 - 1 GAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCGAACUUCGGUUGAGGCCCA-------UUCUGUACCGUGCCGCAGGGCGCGGCACUUUUGUGUGUAUCUUGUGGAUAAACGCUG ..((((((...((((((((((...((((...(((.(.(((....)))).)))...-------.........(((((....)))))))))......))))))))))...))))))..... ( -39.30) >DroEre_CAF1 61310 112 - 1 GAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAUCUUCGGUUGAGGUCCA-------UUCUGUACCGUGCCGCAGGGCGCGGCACUUUUGUGUGUAUCUUGUGGAUAAACGCUG ..((((((...((((((((((...((((...(((.(((.(....)))).)))...-------.........(((((....)))))))))......))))))))))...))))))..... ( -37.40) >DroYak_CAF1 82366 112 - 1 GAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAACUUCGGUUGAGCUCCA-------UUCUGUACCGUGCCGCAGGGCGCGGCACUUUUGUGUGUAUCUUGUGGAUAAACGCUG ..((((((...((((((((((...((((.(.((((..(((....)))..((..((-------(........)))..)).))))).))))......))))))))))...))))))..... ( -35.80) >DroAna_CAF1 49675 117 - 1 GAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCA-GCCCCGGCUGAGGUCCAUGCUGUAUUCUGUAUCUUGCCGCAGGGCGCGGCACUUUU-UGUGUAUCUUAUGGAUAAACGCUG ..((((((.((((((((((((((.((((.(.((((((-((....)))).(((..((((........))))...)))...))))).))))...))-)))))))))).))))))))..... ( -44.00) >consensus GAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAACUUCGGUUGAGGUCCA_______UUCUGUACCGUGCCGCAGGGCGCGGCACUUUUGUGUGUAUCUUGUGGAUAAACGCUG ..((((((...((((((((((...((((...(((.((.((....)))).)))...................(((((....)))))))))......))))))))))...))))))..... (-35.22 = -35.13 + -0.08)

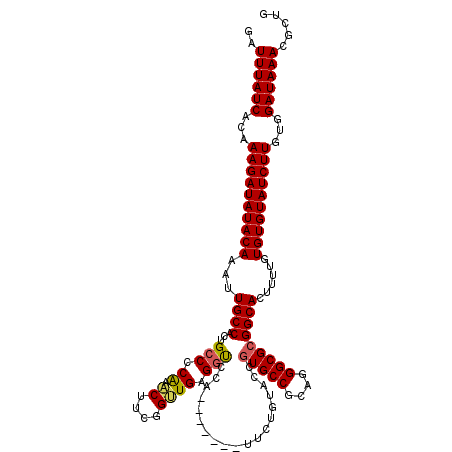

| Location | 17,262,991 – 17,263,103 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 94.05 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -23.97 |
| Energy contribution | -23.72 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17262991 112 - 27905053 AGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAACUUCGGUUGAGGUCCA-------UUCUCAACCGUGCCGCA .((((((...(((...((((......)))).......((((...((((.....))))..))))..)))..............(((((((((....-------.))))))))))))))). ( -32.10) >DroSec_CAF1 59579 112 - 1 AGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCGAACUUCGGUUGAGGUCCA-------UUCUGUACCGUGCCGCA .(((((..((((.(((.((.........((((((.........)))))).....................((..((.(((....)))..))..))-------.)).)))))))))))). ( -25.70) >DroSim_CAF1 62350 112 - 1 AGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCGAACUUCGGUUGAGGCCCA-------UUCUGUACCGUGCCGCA .(((((..((((.(((.((.........((((((.........))))))......................(((.(.(((....)))).)))...-------.)).)))))))))))). ( -27.20) >DroEre_CAF1 61350 112 - 1 AGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAUCUUCGGUUGAGGUCCA-------UUCUGUACCGUGCCGCA .(((((..((((.(((.((.........((((((.........))))))......................(((.(((.(....)))).)))...-------.)).)))))))))))). ( -25.30) >DroYak_CAF1 82406 112 - 1 AGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAACUUCGGUUGAGCUCCA-------UUCUGUACCGUGCCGCA .(((((..((((.(((.((.........((((((.........))))))......................((....(((....)))..))....-------.)).)))))))))))). ( -23.70) >DroAna_CAF1 49714 118 - 1 AGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCA-GCCCCGGCUGAGGUCCAUGCUGUAUUCUGUAUCUUGCCGCA .(((((...(((...)))..........((((((.........))))))..((((((((......((....(((.((-((....)))).)))....)).......))))))))))))). ( -31.72) >consensus AGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAACUUCGGUUGAGGUCCA_______UUCUGUACCGUGCCGCA .((((((...(((...((((......)))).......((((...((((.....))))..))))..)))...(((.((.((....)))).)))....................)))))). (-23.97 = -23.72 + -0.25)



| Location | 17,263,023 – 17,263,143 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -18.20 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.45 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17263023 120 - 27905053 CACAACAAAAAGCCGACAAAUUUCGCCCCAAACUCUUCCAAGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAAC ...........(((((......)))...............((((((((.(((...)))..........((((((.........))))))..............))))))))))....... ( -17.80) >DroSec_CAF1 59611 120 - 1 CACAACAAAAAGCCGACAAAUUUCGCCCCAAACGCUUCCAAGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCGAAC .........(((((((......)))........))))...((((((((.(((...)))..........((((((.........))))))..............))))))))......... ( -18.10) >DroSim_CAF1 62382 120 - 1 CACAACAAAAAGCCGACAAAUUUCGCCCCAAACGCUUCCAAGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCGAAC .........(((((((......)))........))))...((((((((.(((...)))..........((((((.........))))))..............))))))))......... ( -18.10) >DroEre_CAF1 61382 120 - 1 CACAACAAAAAGCCGACAAAUUUCGCCCCAAACGCUUCCAAGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAUC .........(((((((......)))........))))...((((((((.(((...)))..........((((((.........))))))..............))))))))......... ( -18.10) >DroYak_CAF1 82438 120 - 1 CACAACAAAAAGCCGACAAAUUUCGCCCCAAACGCUUCCAAGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAAC .........(((((((......)))........))))...((((((((.(((...)))..........((((((.........))))))..............))))))))......... ( -18.10) >DroAna_CAF1 49753 119 - 1 CACAACAAAAAGCCGACAAAUUUUGCCCCAAACGCUUCCAAGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCA-GC .................................(((....((((((((.(((...)))..........((((((.........))))))..............)))))))).....)-)) ( -19.00) >consensus CACAACAAAAAGCCGACAAAUUUCGCCCCAAACGCUUCCAAGUGGCAAACGGCUACCGAUUUUAUUAUUGAUAAUGUUUUGAUUUAUCACAAAGAUAUACAAAUUGCCACUGCCCCAAAC ...........(((((......)))...............((((((((.(((...)))..........((((((.........))))))..............))))))))))....... (-17.59 = -17.45 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:24 2006