
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,254,957 – 17,255,152 |
| Length | 195 |
| Max. P | 0.947219 |

| Location | 17,254,957 – 17,255,073 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -30.67 |
| Energy contribution | -30.33 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17254957 116 - 27905053 UACACAGAUACACGACGGCUGCAGAGGUGUGCGAAAGCGACGCACUGAAGGUGUUGCCUGCACGCUUUUGAUGAGCACUAAUUGAGCUCAAGUUGCAUUUUUCUUCUCUGCCCAUU---- ................((..(((((((((((((...(((((((.(....)))))))).)))))))......(((((.(.....).)))))...............))))))))...---- ( -36.60) >DroSec_CAF1 51505 116 - 1 UACACAGAUACUCGCCGGCUGAAAAGGUGUGCGAAAGCGACGCACUGAAGGUGUUGCCUACACGCUUUUGAUGAGCACUAAUUGAGCUCAAGUUGCAUUUUUCUUCUCUGCCCAUU---- ....((((...((((((.((....)).)).))))..(((((...(.((((((((.....)))).)))).).(((((.(.....).))))).)))))..........))))......---- ( -33.10) >DroSim_CAF1 52104 116 - 1 UACACAGAUACUCGCCGGCUGAAAAGGUGUGCGAAAGCGACGCACUGAAGGUGUUGCCUGCACGCUUUUGAUGAGCACUAAUUGAGCUCAAGUUGCAUUUUUCUUCUCUGCCCAUU---- ................(((.(((((((((((((...(((((((.(....)))))))).)))))))))))..(((((.(.....).)))))................)).)))....---- ( -34.60) >DroEre_CAF1 53479 116 - 1 UACACAGAUACUCGACGGCUGAAGAGGUGUGCGAAAGCGACGCACUGAAGGUGUUGCCUGCACGCUUUUGAUGAGCACUAAUUGAGCUCAAGUUGCAUUUUUCUUCUCUGCCCAUU---- ................(((.(((((((((((((...(((((((.(....)))))))).)))))))).....(((((.(.....).)))))...........)))))...)))....---- ( -36.20) >DroYak_CAF1 74419 116 - 1 UACACAGAUACUCACCAGCUGAGAAGGUGUGCGAAAGCGACGCACUGAAGGUGUUGCCUGCACGCUUUUGAUGAGCACUAAUUGAGCUCAAGUUGCAUUUUUCUUCUCUGCCCAUU---- .................((.(((((((((((((...(((((((.(....)))))))).)))))))......(((((.(.....).)))))............)))))).)).....---- ( -35.90) >DroAna_CAF1 42244 111 - 1 UACUCGGAUACAC---------AGAGGUGUGCGAAAGCAACGCACUGAAGGUGUUACCUGCACGCUUUUGAUGAGCACUAAUUGAGCUCAAGUUGCAUUUUUCUUUUCUGCCCAAAGGAG ...(((.((((..---------....)))).)))..(((((...(.((((((((.....)))).)))).).(((((.(.....).))))).))))).....((((((......)))))). ( -29.60) >consensus UACACAGAUACUCGCCGGCUGAAAAGGUGUGCGAAAGCGACGCACUGAAGGUGUUGCCUGCACGCUUUUGAUGAGCACUAAUUGAGCUCAAGUUGCAUUUUUCUUCUCUGCCCAUU____ ....((((..............(((((((((((...(((((((.(....)))))))).)))))))))))..(((((.(.....).)))))................)))).......... (-30.67 = -30.33 + -0.33)


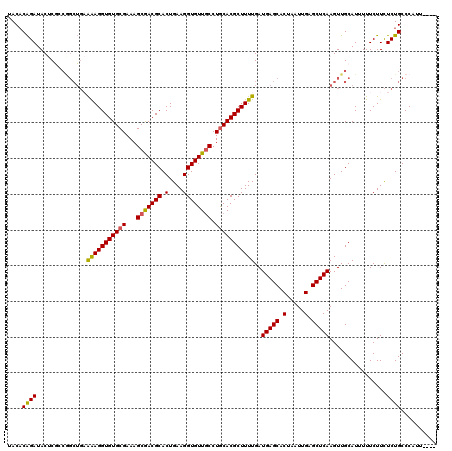
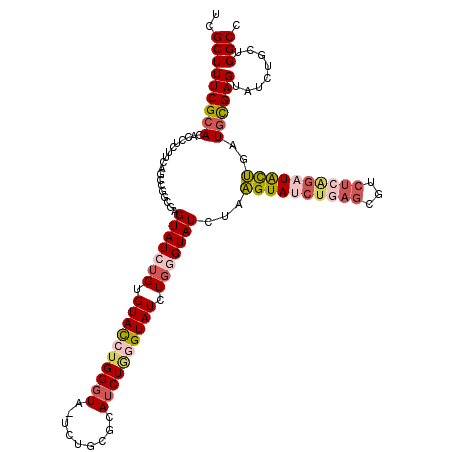
| Location | 17,255,033 – 17,255,152 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -20.32 |
| Energy contribution | -22.68 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17255033 119 + 27905053 UCGCUUUCGCACACCUCUGCAGCCGUCGUGUAUCUGUGUACCUGGGUA-UCUGGGCAUCUGGGUAUCUGGGUAUCUAAGUAUCUGAGCGUCUCAGAUACUGAUGCGAGUAUAUGCUGGCC ........(((......))).((((.((((((..((((((((..((((-((..(....)..))))))..)))))...((((((((((...))))))))))...)))..)))))).)))). ( -48.00) >DroSec_CAF1 51581 119 + 1 UCGCUUUCGCACACCUUUUCAGCCGGCGAGUAUCUGUGUACCUGGGUA-UCUGCGCAUCUGGGUAUCUGGGUAUCUAAGUAUCUGAGCGUCUCAGAUACUGAUGCGAGAAUCUGCUGGCC ..((....))...........(((((((.(((((...(((((..((((-(((........)))))))..)))))...((((((((((...)))))))))))))))(.....)))))))). ( -45.60) >DroSim_CAF1 52180 119 + 1 UCGCUUUCGCACACCUUUUCAGCCGGCGAGUAUCUGUGUACCUGGGUA-UCUGCGCAUCUGGGUAUCUGGGUAUCUAAGUAUCUGAGCGUCUCAGAUACUGAUGCGAGUAUCUGCUGGCC ..((....))...........(((((((..((..((((((((..((((-(((........)))))))..)))))...((((((((((...))))))))))...)))..))..))))))). ( -46.90) >DroEre_CAF1 53555 98 + 1 UCGCUUUCGCACACCUCUUCAGCCGUCGAGUAUCUGUGUACCUGGGUA-UCUUAGUAUCUAAGUAUCUGAGUAU----------------CUCGGU-----GUGCGAGUAUCUGCUGGCC ..((....))...........((((.((..((..((..((((.(((((-.((((((((....))).))))))))----------------)).)))-----)..))..))..)).)))). ( -28.70) >DroAna_CAF1 42324 106 + 1 UUGCUUUCGCACACCUCU---------GUGUAUCCGAGUAUCUGGCUGGUC--UGCAUCU--GUAUCUAAGUAUCUGUGUAUCUUAGCAUCUCGG-UGUGUAUCUGAGCAUCUCCAGGCC .(((((..(((((((...---------(......)(((......(((((..--(((((..--((((....))))..)))))..)))))..)))))-)))))....))))).......... ( -26.40) >consensus UCGCUUUCGCACACCUCUUCAGCCGGCGAGUAUCUGUGUACCUGGGUA_UCUGCGCAUCUGGGUAUCUGGGUAUCUAAGUAUCUGAGCGUCUCAGAUACUGAUGCGAGUAUCUGCUGGCC ..(((((((((..................(((((((.((((((((((.........)))))))))).)))))))...((((((((((...))))))))))..))))))........))). (-20.32 = -22.68 + 2.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:17 2006