
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,249,458 – 17,249,725 |
| Length | 267 |
| Max. P | 0.992290 |

| Location | 17,249,458 – 17,249,576 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.60 |
| Mean single sequence MFE | -17.66 |
| Consensus MFE | -14.46 |
| Energy contribution | -15.86 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17249458 118 - 27905053 UAC--UAUACACUGUGCCAUACGACGGCAGCCACAAUAACAAUAAUAAUAAUGCAUUCAUCACUUUAUGACGGCCCCACAUACAAACUUGGCCUAGACAAUAGUGUCAACCAUAAAGAGA ...--.........((((.......))))..............................((.(((((((..((((..............))))..((((....))))...))))))).)) ( -20.64) >DroSec_CAF1 45884 115 - 1 -----CAAACACUGUGCCAUACAACGGCAGCCACAAUAACAAUAAUAAUAAUGCAUUCAUCACUUUAUGACGGCCCCACAUACAAACUUGGCCUAGACAAUAGUGUCAACCAUAAAGAGA -----.........((((.......))))..............................((.(((((((..((((..............))))..((((....))))...))))))).)) ( -20.64) >DroEre_CAF1 47700 120 - 1 UACCACAUACACUGUGCCAUACAACGGCAGCCACAAUAACAAUAAUAAUAAUGCAUUCAUCACCUUAUGACGGCCUCACAUACAAACUUGGCCUAGACAAUAGUGUCAACCAUAAAGAGA ..............((((.......))))..............................((.(.(((((..((((..............))))..((((....))))...))))).).)) ( -17.74) >DroYak_CAF1 68705 120 - 1 UACUACAUACACUGUACCAUACAACGGCAGCCACAAUAACAAUAAUAAUAAUGCAUUCAUCACUUUAUGACGGCCCCACAUACAAACUUGGCCUAGACAAUAGUGUCAACCAUAAAGAGA ...........((((...........)))).............................((.(((((((..((((..............))))..((((....))))...))))))).)) ( -17.34) >DroAna_CAF1 37495 107 - 1 --------ACA-----CAGCACAACACCGGCCACAAUAACAAUAAUAAUAAUGCAUUCAUCACUUUAUGACAGCCCCACAUACAAACUUGGCCUAGACAAUAGUGUCAACCAUAAAGAGA --------...-----...........................................((.(((((((...(((..............)))...((((....))))...))))))).)) ( -11.94) >consensus UAC__CAUACACUGUGCCAUACAACGGCAGCCACAAUAACAAUAAUAAUAAUGCAUUCAUCACUUUAUGACGGCCCCACAUACAAACUUGGCCUAGACAAUAGUGUCAACCAUAAAGAGA ..............((((.......))))..............................((.(((((((..((((..............))))..((((....))))...))))))).)) (-14.46 = -15.86 + 1.40)



| Location | 17,249,498 – 17,249,614 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -28.46 |
| Energy contribution | -28.68 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17249498 116 + 27905053 UGUGGGGCCGUCAUAAAGUGAUGAAUGCAUUAUUAUUAUUGUUAUUGUGGCUGCCGUCGUAUGGCACAGUGUAUA--GUAUAGUAUAGUGUAUAGUGUACAUUGUGUAUUGCUGGCUG .....(((.(((((((((((((((........))))))))....))))))).)))((((((..((((((((((((--.((((........)))).))))))))))))..))).))).. ( -38.00) >DroSec_CAF1 45924 106 + 1 UGUGGGGCCGUCAUAAAGUGAUGAAUGCAUUAUUAUUAUUGUUAUUGUGGCUGCCGUUGUAUGGCACAGUGUUUG-----UA-------GUAUAGUGUACAUUGUGUAUUGCUGGCUG .....(((((.((....(..(((..((((((((.(((((...(((((((.((((....))).).)))))))...)-----))-------)))))))))))))..)....)).))))). ( -31.00) >DroEre_CAF1 47740 104 + 1 UGUGAGGCCGUCAUAAGGUGAUGAAUGCAUUAUUAUUAUUGUUAUUGUGGCUGCCGUUGUAUGGCACAGUGUAUGUGGUAUAG--------------UACAUUGUGUAUUGCUGGCUG .....(((((..((((((((((((........)))))))).))))..)))))((((..(((..(((((((((((........)--------------))))))))))..))))))).. ( -32.00) >consensus UGUGGGGCCGUCAUAAAGUGAUGAAUGCAUUAUUAUUAUUGUUAUUGUGGCUGCCGUUGUAUGGCACAGUGUAUG__GUAUAG______GUAUAGUGUACAUUGUGUAUUGCUGGCUG .....(((.(((((((((((((((........))))))))....))))))).)))((((((..(((((((((((......................)))))))))))..))).))).. (-28.46 = -28.68 + 0.23)

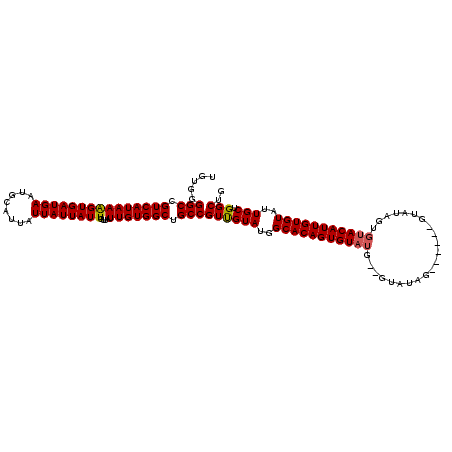
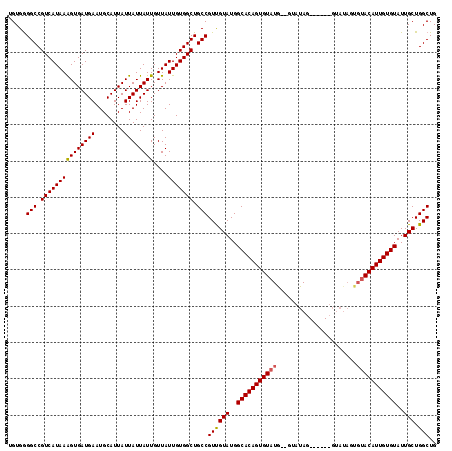
| Location | 17,249,498 – 17,249,614 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -15.43 |
| Consensus MFE | -13.63 |
| Energy contribution | -13.63 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17249498 116 - 27905053 CAGCCAGCAAUACACAAUGUACACUAUACACUAUACUAUAC--UAUACACUGUGCCAUACGACGGCAGCCACAAUAACAAUAAUAAUAAUGCAUUCAUCACUUUAUGACGGCCCCACA ..(((.(((....(((.((((.(.((((........)))).--).)))).)))(((.......))).......................)))..((((......)))).)))...... ( -16.50) >DroSec_CAF1 45924 106 - 1 CAGCCAGCAAUACACAAUGUACACUAUAC-------UA-----CAAACACUGUGCCAUACAACGGCAGCCACAAUAACAAUAAUAAUAAUGCAUUCAUCACUUUAUGACGGCCCCACA ..(((.(((........((((........-------))-----)).......((((.......))))......................)))..((((......)))).)))...... ( -14.60) >DroEre_CAF1 47740 104 - 1 CAGCCAGCAAUACACAAUGUA--------------CUAUACCACAUACACUGUGCCAUACAACGGCAGCCACAAUAACAAUAAUAAUAAUGCAUUCAUCACCUUAUGACGGCCUCACA ..(((.(((....(((.((((--------------..........)))).)))(((.......))).......................)))..((((......)))).)))...... ( -15.20) >consensus CAGCCAGCAAUACACAAUGUACACUAUAC______CUAUAC__CAUACACUGUGCCAUACAACGGCAGCCACAAUAACAAUAAUAAUAAUGCAUUCAUCACUUUAUGACGGCCCCACA ..(((.(((.(((.....)))...............................((((.......))))......................)))..((((......)))).)))...... (-13.63 = -13.63 + 0.00)



| Location | 17,249,538 – 17,249,654 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.71 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.78 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17249538 116 + 27905053 GUUAUUGUGGCUGCCGUCGUAUGGCACAGUGUAUA--GUAUAGUAUAGUGUAUAGUGUACAUUGUGUAUUGCUGGCUGGGCCUUUGUUGAAAAGCGUUAAUUGAGUGUUGCGAAUGUG ....(((..((.(((((((((..((((((((((((--.((((........)))).))))))))))))..))).)))..)))....(((....)))...........))..)))..... ( -34.40) >DroSec_CAF1 45964 106 + 1 GUUAUUGUGGCUGCCGUUGUAUGGCACAGUGUUUG-----UA-------GUAUAGUGUACAUUGUGUAUUGCUGGCUGGGCCUUUGUUGAAAAGCGUUAAUUGAGUGUUGCGAAUGUG ....((((((((((((..(((..(((((((((...-----..-------.........)))))))))..)))))))..))))...(((....)))..............))))..... ( -25.44) >DroEre_CAF1 47780 104 + 1 GUUAUUGUGGCUGCCGUUGUAUGGCACAGUGUAUGUGGUAUAG--------------UACAUUGUGUAUUGCUGGCUGGGCCUUUGUUGAAAAGCGUUAAUUGAGUGUUGCGAAUGUG ....((((((((((((..(((..(((((((((((........)--------------))))))))))..)))))))..))))...(((....)))..............))))..... ( -29.40) >consensus GUUAUUGUGGCUGCCGUUGUAUGGCACAGUGUAUG__GUAUAG______GUAUAGUGUACAUUGUGUAUUGCUGGCUGGGCCUUUGUUGAAAAGCGUUAAUUGAGUGUUGCGAAUGUG ....(((..((.((((..(((..(((((((((((......................)))))))))))..)))))))..(((((((.....)))).)))........))..)))..... (-25.12 = -25.78 + 0.67)



| Location | 17,249,538 – 17,249,654 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.71 |
| Mean single sequence MFE | -13.81 |
| Consensus MFE | -12.02 |
| Energy contribution | -12.35 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17249538 116 - 27905053 CACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCCCAGCCAGCAAUACACAAUGUACACUAUACACUAUACUAUAC--UAUACACUGUGCCAUACGACGGCAGCCACAAUAAC .......((..............((((((....))))))...(((..(....((((.((((.(.((((........)))).--).)))).))))......)..))).))......... ( -15.10) >DroSec_CAF1 45964 106 - 1 CACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCCCAGCCAGCAAUACACAAUGUACACUAUAC-------UA-----CAAACACUGUGCCAUACAACGGCAGCCACAAUAAC .......((..............((((((....))))))...(((.......((((.(((.........-------..-----...))).)))).........))).))......... ( -12.53) >DroEre_CAF1 47780 104 - 1 CACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCCCAGCCAGCAAUACACAAUGUA--------------CUAUACCACAUACACUGUGCCAUACAACGGCAGCCACAAUAAC .......((..............((((((....))))))...(((.......((((.((((--------------..........)))).)))).........))).))......... ( -13.79) >consensus CACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCCCAGCCAGCAAUACACAAUGUACACUAUAC______CUAUAC__CAUACACUGUGCCAUACAACGGCAGCCACAAUAAC .......((..............((((((....))))))...(((.......((((.((((........................)))).)))).........))).))......... (-12.02 = -12.35 + 0.33)

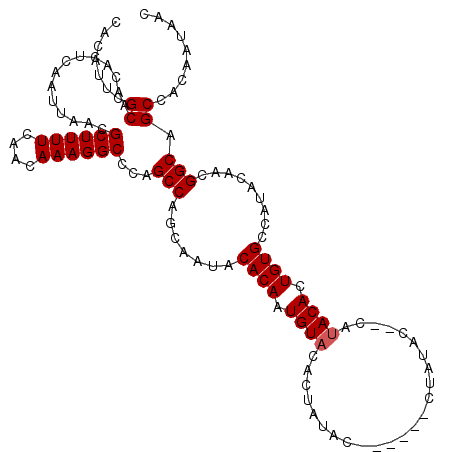

| Location | 17,249,576 – 17,249,694 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.42 |
| Mean single sequence MFE | -19.84 |
| Consensus MFE | -18.33 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17249576 118 - 27905053 GGCAGCAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCCCAGCCAGCAAUACACAAUGUACACUAUACACUAU--ACUA (((.(((((.((((.................)))).)))))......................((((((....))))))...)))............((((.....))))....--.... ( -20.13) >DroSec_CAF1 45999 111 - 1 GGCAGCAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCCCAGCCAGCAAUACACAAUGUACACUAUAC---------UA (((.(((((.((((.................)))).)))))......................((((((....))))))...)))........................---------.. ( -19.73) >DroEre_CAF1 47820 104 - 1 GGCAGCAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCCCAGCCAGCAAUACACAAUGUA----------------CUA (((.(((((.((((.................)))).)))))......................((((((....))))))...)))................----------------... ( -19.73) >DroYak_CAF1 68825 120 - 1 GGCAGCAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCCCAGCCAGCAAUACACAAUGUACACUAUACACUAUCCACUA (((.(((((.((((.................)))).)))))......................((((((....))))))...)))............((((.....)))).......... ( -20.13) >DroAna_CAF1 37602 97 - 1 GGCAACAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCCCGGCCAGCAAAACGGCA----------------------- .((......(((..(((............(((((..(((((......)))))...)))))...((((((....))))))..)))..))).....)).----------------------- ( -19.50) >consensus GGCAGCAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCCCAGCCAGCAAUACACAAUGUACACUAUAC________CUA (((.(((((.((((.................)))).)))))......................((((((....))))))...)))................................... (-18.33 = -18.53 + 0.20)


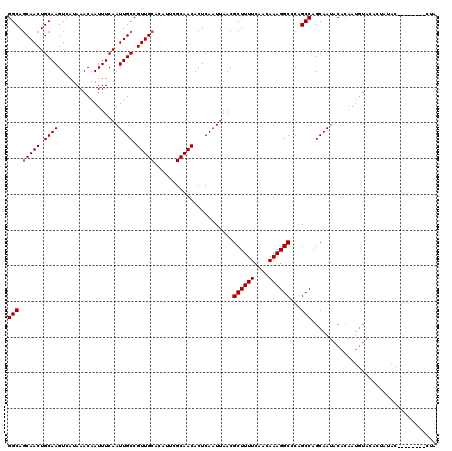
| Location | 17,249,614 – 17,249,725 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.26 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -21.74 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17249614 111 - 27905053 CAUUACA-------CAUCAUUGAGCAUGAGCUAAGGCU--GGCAGCAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCC .......-------....((((((..........((((--.((((...)))).))))...................(((((......))))).))))))....((((((....)))))). ( -23.80) >DroSec_CAF1 46030 111 - 1 CGUUACA-------CAUUAUUGAGCAUGAGCUACGGCU--GGCAGCAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCC .((((..-------....((((((....(((....)))--((((((....))..((.....)).........))))(((((......))))).))))))))))((((((....)))))). ( -27.10) >DroEre_CAF1 47844 118 - 1 CAUUACACAUUGCACAUUAUUGGGCAUGAGCUACGGCA--GGCAGCAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCC ..........(((.((....)).)))((((....((((--(((((...))))..((.....))........)))))(((((......))))).))))......((((((....)))))). ( -26.40) >DroYak_CAF1 68865 118 - 1 CAUUACACAUUACACAUUAUUGAGCAUGAGCUACGGCA--GGCAGCAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCC ..................((((((.....((....)).--((((((....))..((.....)).........))))(((((......))))).))))))....((((((....)))))). ( -25.30) >DroAna_CAF1 37619 111 - 1 CAUUAC---------AUUAUUGAACAUGAGCCCGGGCCGCGGCAACAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCC ......---------...........((((....(((.((((......))))..((.....))..........)))(((((......))))).))))......((((((....)))))). ( -21.60) >consensus CAUUACA_______CAUUAUUGAGCAUGAGCUACGGCA__GGCAGCAACUGCAAGUCAUAAACAAUUUCAAUUGCCGUUGCACAUUCGCAACACUCAAUUAACGCUUUUCAACAAAGGCC ..................((((((.....((....))...((((((....))..((.....)).........))))(((((......))))).))))))....((((((....)))))). (-21.74 = -21.98 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:09 2006