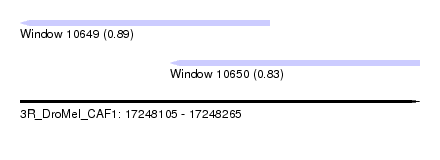
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,248,105 – 17,248,265 |
| Length | 160 |
| Max. P | 0.886846 |

| Location | 17,248,105 – 17,248,205 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -23.08 |
| Energy contribution | -22.68 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
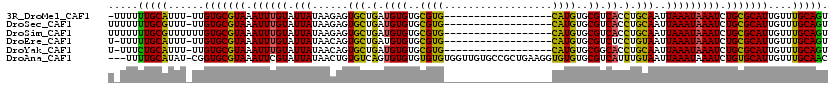
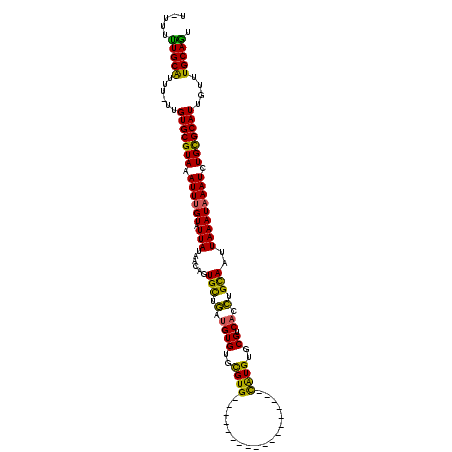
>3R_DroMel_CAF1 17248105 100 - 27905053 CGUCACCUGCAAUUAAAUAAAUCUGCGCAUUGUUUGCAGUGCCGUUAAAGCGGCAACCAUAAAUGGGACAGUAGU--------AUACGCAACAGCGGGAGUU-------ACCCUA .....(((((............((((.((((...((...((((((....))))))..))..)))).).)))..((--------....))....)))))....-------...... ( -25.50) >DroSec_CAF1 44531 100 - 1 CGUCACCUGCAAUUAAAUAAAUCUGCGCAUUGUUUGCAGUGCCGUUAAAGCGGCAACCAUAAAUGGGACAGUAGU--------AUUUGCAACAGCGGGCGUU-------ACACUA ((((..(((....((((((...((((.((((...((...((((((....))))))..))..)))).).)))...)--------)))))...)))..))))..-------...... ( -29.30) >DroSim_CAF1 45230 100 - 1 CGUCACCUGCAAUUAAAUAAAUCUGCGCAUUGUUUGCAGUGCCGUUAAAGCGGCAACCAUAAAUGGGACAGUAGU--------AUUUGCAGCAGCGGGCGUU-------ACACUA ((((..((((...((((((...((((.((((...((...((((((....))))))..))..)))).).)))...)--------)))))..))))..))))..-------...... ( -33.10) >DroEre_CAF1 45294 97 - 1 CGUCUCCUGUAAUUAAAUAAAUCUGCGCAUUGUUUGCAGUGCCGUUAAAGCGGCAACCAUAAACGGGACAGU-----------AUAUGCAACAGCAGGCGAU-------CUACUA ((((((((((............(((((.......)))))((((((....)))))).......))))))....-----------...(((....)))))))..-------...... ( -26.90) >DroYak_CAF1 67311 115 - 1 CGGCACCUGCAAUUAAAUAAAUCUGCGCAUUGUUUGCAGUGCCGUUAAAGCGGCAACCAUAAAUGGGACAGUAGUACAUACAUAUAUGCAACAGCAGGCGGUAUACUAUGUACUA ..((....))............((((.((((...((...((((((....))))))..))..)))).).)))(((((((((.((((.(((....)))....))))..))))))))) ( -30.90) >consensus CGUCACCUGCAAUUAAAUAAAUCUGCGCAUUGUUUGCAGUGCCGUUAAAGCGGCAACCAUAAAUGGGACAGUAGU________AUAUGCAACAGCGGGCGUU_______ACACUA ...(.(((((............(((((.......)))))((((((....)))))).((......))...........................))))).)............... (-23.08 = -22.68 + -0.40)

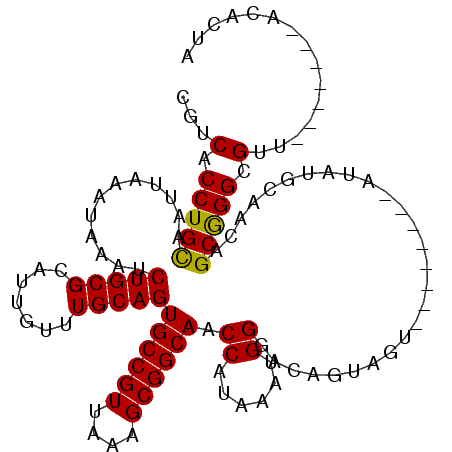
| Location | 17,248,165 – 17,248,265 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.93 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -22.64 |
| Energy contribution | -21.29 |
| Covariance contribution | -1.35 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17248165 100 - 27905053 -UUUUUUGCAUUU-UUGUGCGUAAAUUUGUAUUAUAAGAGUGCUGAUGUGUGCGUG------------------CAUGUGCGUCACCUGCAAUUAAAUAAAUCUGCGCAUUGUUUGCAGU -....(((((...-..(((((((.((((((.........(..(......)..).((------------------((.(((...))).)))).....)))))).)))))))....))))). ( -26.90) >DroSec_CAF1 44591 101 - 1 UUUUUUUGCGUUU-UUGUGCGUAAAUUUGUAUUAUAAGAGUGCUGAUGUGUGCGUG------------------CAUGUGCGUCACCUGCAAUUAAAUAAAUCUGCGCAUUGUUUGCAGU .....(((((...-..(((((((.((((((.........(..(......)..).((------------------((.(((...))).)))).....)))))).)))))))....))))). ( -26.20) >DroSim_CAF1 45290 102 - 1 UUUUUUUGCGUUUUUUGUGCGUAAAUUUGUAUUAUAAGAGUGCUGAUGUGUGCGUG------------------CAUGUGCGUCACCUGCAAUUAAAUAAAUCUGCGCAUUGUUUGCAGU .....(((((..(...(((((((.((((((.........(..(......)..).((------------------((.(((...))).)))).....)))))).))))))).)..))))). ( -25.80) >DroEre_CAF1 45351 100 - 1 U-UUUUUGCAUUU-UUGUGCGUAAAUUUGUAUUAUAACAGUGCUGAUGUGUGCGUG------------------CAUGUGCGUCUCCUGUAAUUAAAUAAAUCUGCGCAUUGUUUGCAGU .-...(((((...-..(((((((.((((((...((.((((.(..((((((((....------------------)))..))))).))))).))...)))))).)))))))....))))). ( -27.30) >DroYak_CAF1 67386 100 - 1 U-UUUCUGCAUUU-UUGUGCGUAAAUUUGUAUUAUAACAGUGCUGAUGUGUGCGUG------------------CAUGUGCGGCACCUGCAAUUAAAUAAAUCUGCGCAUUGUUUGCAGU .-...(((((...-..(((((((.((((((.(((...((((((((((((((....)------------------))))).))))).)))....))))))))).)))))))....))))). ( -32.90) >DroAna_CAF1 36380 116 - 1 ---UUUUGCAUAU-CGGUGCGUAAAUUCGUAUUAUAACUGUGUCAGUGUGUGUGUGUGGUUGUGCCGCUGAAGGUGUGUGCGUCAUUUGUAAUUAAAUAAAUCUGUGCAUUGUUUGCAAC ---..(((((...-(((((((......(((((....(((...((((((.(..((......))..))))))).)))..)))))..((((((......))))))...)))))))..))))). ( -30.00) >consensus U_UUUUUGCAUUU_UUGUGCGUAAAUUUGUAUUAUAACAGUGCUGAUGUGUGCGUG__________________CAUGUGCGUCACCUGCAAUUAAAUAAAUCUGCGCAUUGUUUGCAGU .....(((((......(((((((.((((((.(((......(((.(.((((..((((..................))))..)).)).).)))..))))))))).)))))))....))))). (-22.64 = -21.29 + -1.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:03 2006