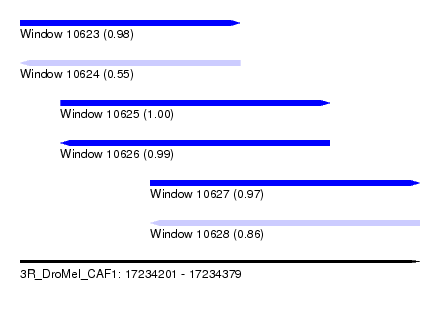
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,234,201 – 17,234,379 |
| Length | 178 |
| Max. P | 0.999440 |

| Location | 17,234,201 – 17,234,299 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -24.14 |
| Energy contribution | -25.14 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17234201 98 + 27905053 ----------------------AUUGAAAUUAAAAGAAAUCGGUAAAUAAUAGCCUCACCACUAGGUGGAGCUCGAGUGCUAUCUCUAAAGCAGUUCUCUUGCUGCUUGCCGAAGCAGUU ----------------------.................(((((.......(((..((((....))))..))).(((......)))..(((((((......))))))))))))....... ( -28.00) >DroSec_CAF1 29852 111 + 1 GUUGAGUGCCGUU---------AUUCAAAUUAAAAGAAAUCGGUAAAUAAUAGCUUCACCACUAGGUGGAGCUCGAGUGCUAUCUCUGAAGCAGUUCUGUGGCUGCUUGCCGAAGCAGUU .(((((((....)---------))))))...........(((((.......(((((((((....))))))))).(((......)))..(((((((......))))))))))))....... ( -34.80) >DroSim_CAF1 31024 111 + 1 GUUGCGUGCCGCU---------AUUCAAAUUAAAAGAAAUCGGUAAACAAUUGCUUCACCACUAGGUGGAGCUCGAGUGCUAUCUCUGAAGCAGUUCUCUGGCUGCUUGCCGAAGCAGUU ((((..(((((..---------.(((.........)))..)))))..)))).((((((((....))))))))..((.((((...((.((((((((......))))))).).)))))).)) ( -34.80) >DroEre_CAF1 31662 118 + 1 ACUUAUUGU--UAAUUUAAAACUUUCAGAUUAAAAGUAAUCAGUUAGUAAUGGCAUAACCGCUAGGUGGAGCACGAGUGCGAUCUGUAAAGCAGUUCACUCGCUGCUAGCCGAAGCAGUC ....(((((--(........(((..(.(((((....))))).)..)))..((((....((((...))))((((((((((....(((.....)))..)))))).)))).)))).)))))). ( -32.10) >DroYak_CAF1 50025 118 + 1 GUUCAUUGU--UCAUUUAAAACUUGUAUAUAAAAAGAAAUCUGUAAGUAAUGGCAUCACCGCUAGGUGGUGCUCGAGUGCGAUCUCUAAAGCAGUUUUCUUGCUGCUCGCCGAAGCAGUC .........--......................((((((.((((.......(((((((((....))))))))).(((......)))....)))).))))))((((((......)))))). ( -32.70) >consensus GUUGAGUGC__UU_________AUUCAAAUUAAAAGAAAUCGGUAAAUAAUAGCAUCACCACUAGGUGGAGCUCGAGUGCUAUCUCUAAAGCAGUUCUCUGGCUGCUUGCCGAAGCAGUU .......................................(((((((......((((((((....))))))))..(((......)))....(((((......))))))))))))....... (-24.14 = -25.14 + 1.00)



| Location | 17,234,201 – 17,234,299 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -19.30 |
| Energy contribution | -19.66 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17234201 98 - 27905053 AACUGCUUCGGCAAGCAGCAAGAGAACUGCUUUAGAGAUAGCACUCGAGCUCCACCUAGUGGUGAGGCUAUUAUUUACCGAUUUCUUUUAAUUUCAAU---------------------- ..((((((....)))))).(((((((.((((........))))....(((..((((....))))..))).............))))))).........---------------------- ( -24.90) >DroSec_CAF1 29852 111 - 1 AACUGCUUCGGCAAGCAGCCACAGAACUGCUUCAGAGAUAGCACUCGAGCUCCACCUAGUGGUGAAGCUAUUAUUUACCGAUUUCUUUUAAUUUGAAU---------AACGGCACUCAAC ....((....))((((((........))))))..(((......)))(((...((((....))))..((..(((((((..((((......)))))))))---------))..)).)))... ( -28.20) >DroSim_CAF1 31024 111 - 1 AACUGCUUCGGCAAGCAGCCAGAGAACUGCUUCAGAGAUAGCACUCGAGCUCCACCUAGUGGUGAAGCAAUUGUUUACCGAUUUCUUUUAAUUUGAAU---------AGCGGCACGCAAC ..(((((((((.((((((...(((...((((........)))))))..(((.((((....)))).)))..)))))).)))).(((.........))).---------)))))........ ( -29.80) >DroEre_CAF1 31662 118 - 1 GACUGCUUCGGCUAGCAGCGAGUGAACUGCUUUACAGAUCGCACUCGUGCUCCACCUAGCGGUUAUGCCAUUACUAACUGAUUACUUUUAAUCUGAAAGUUUUAAAUUA--ACAAUAAGU (((((((..((..((((.((((((..(((.....)))....))))))))))...)).)))))))..............(((((((((((.....))))))....)))))--......... ( -27.90) >DroYak_CAF1 50025 118 - 1 GACUGCUUCGGCGAGCAGCAAGAAAACUGCUUUAGAGAUCGCACUCGAGCACCACCUAGCGGUGAUGCCAUUACUUACAGAUUUCUUUUUAUAUACAAGUUUUAAAUGA--ACAAUGAAC (.((((((....)))))))((((((.(((.....(((......)))(.(((.((((....)))).))))........))).))))))...........((((.....))--))....... ( -26.20) >consensus AACUGCUUCGGCAAGCAGCAAGAGAACUGCUUUAGAGAUAGCACUCGAGCUCCACCUAGUGGUGAAGCCAUUAUUUACCGAUUUCUUUUAAUUUGAAU_________AA__ACAAUCAAC ....((....))((((((........))))))..(((......)))..(((.((((....)))).))).................................................... (-19.30 = -19.66 + 0.36)



| Location | 17,234,219 – 17,234,339 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -39.94 |
| Consensus MFE | -28.48 |
| Energy contribution | -29.88 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17234219 120 + 27905053 CGGUAAAUAAUAGCCUCACCACUAGGUGGAGCUCGAGUGCUAUCUCUAAAGCAGUUCUCUUGCUGCUUGCCGAAGCAGUUGAUGCGCUGCUUGUCGAAGCAGUUGAUGCACUGCUUGCAC ((((.......(((..((((....))))..))).(((......)))..(((((((......)))))))))))(((((((.(...((((((((....)))))).))...)))))))).... ( -43.80) >DroSec_CAF1 29883 96 + 1 CGGUAAAUAAUAGCUUCACCACUAGGUGGAGCUCGAGUGCUAUCUCUGAAGCAGUUCUGUGGCUGCUUGCCGAAGCAGUUGAU------------------------GAACUGCUUGCAU .((((......(((((((((....)))))))))....)))).....(((((((((((...((((((((....))))))))...------------------------))))))))).)). ( -39.90) >DroSim_CAF1 31055 96 + 1 CGGUAAACAAUUGCUUCACCACUAGGUGGAGCUCGAGUGCUAUCUCUGAAGCAGUUCUCUGGCUGCUUGCCGAAGCAGUUGAU------------------------GAACUGCUUGCAU .((((.((....((((((((....))))))))....))..))))..(((((((((((..(((((((((....)))))))))..------------------------))))))))).)). ( -40.40) >DroEre_CAF1 31700 96 + 1 CAGUUAGUAAUGGCAUAACCGCUAGGUGGAGCACGAGUGCGAUCUGUAAAGCAGUUCACUCGCUGCUAGCCGAAGCAGUCGAU------------------------UUACUGCUUGCAU ..((((....))))......((..(((..((((((((((....(((.....)))..)))))).)))).))).(((((((....------------------------..))))))))).. ( -34.40) >DroYak_CAF1 50063 96 + 1 CUGUAAGUAAUGGCAUCACCGCUAGGUGGUGCUCGAGUGCGAUCUCUAAAGCAGUUUUCUUGCUGCUCGCCGAAGCAGUCGAU------------------------UCACUGCUUGCAU .((((((((.((((((((((....))))))))((((.(((..((.....((((((......))))))....)).))).)))).------------------------.)).)))))))). ( -41.20) >consensus CGGUAAAUAAUAGCAUCACCACUAGGUGGAGCUCGAGUGCUAUCUCUAAAGCAGUUCUCUGGCUGCUUGCCGAAGCAGUUGAU________________________GAACUGCUUGCAU ..((((......((((((((....))))))))((((.((((.((....(((((((......)))))))...)))))).))))................................)))).. (-28.48 = -29.88 + 1.40)

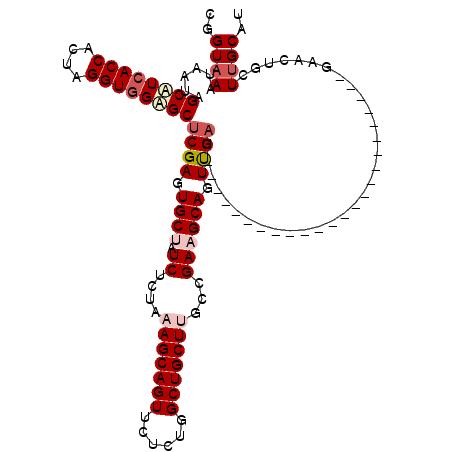
| Location | 17,234,219 – 17,234,339 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -22.95 |
| Energy contribution | -23.11 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17234219 120 - 27905053 GUGCAAGCAGUGCAUCAACUGCUUCGACAAGCAGCGCAUCAACUGCUUCGGCAAGCAGCAAGAGAACUGCUUUAGAGAUAGCACUCGAGCUCCACCUAGUGGUGAGGCUAUUAUUUACCG ((((.((((((......))))))((...((((((..(.((..((((((....))))))...)))..))))))..))....))))...(((..((((....))))..)))........... ( -40.90) >DroSec_CAF1 29883 96 - 1 AUGCAAGCAGUUC------------------------AUCAACUGCUUCGGCAAGCAGCCACAGAACUGCUUCAGAGAUAGCACUCGAGCUCCACCUAGUGGUGAAGCUAUUAUUUACCG .((.(((((((((------------------------.....((((((....)))))).....)))))))))))(((......))).((((.((((....)))).))))........... ( -33.20) >DroSim_CAF1 31055 96 - 1 AUGCAAGCAGUUC------------------------AUCAACUGCUUCGGCAAGCAGCCAGAGAACUGCUUCAGAGAUAGCACUCGAGCUCCACCUAGUGGUGAAGCAAUUGUUUACCG .((.(((((((((------------------------.((..((((((....))))))...)))))))))))))(((......)))..(((.((((....)))).)))............ ( -33.30) >DroEre_CAF1 31700 96 - 1 AUGCAAGCAGUAA------------------------AUCGACUGCUUCGGCUAGCAGCGAGUGAACUGCUUUACAGAUCGCACUCGUGCUCCACCUAGCGGUUAUGCCAUUACUAACUG ..(.(((((((..------------------------....))))))))(((((((.(((((((..(((.....)))....)))))))(((......))).)))).)))........... ( -30.60) >DroYak_CAF1 50063 96 - 1 AUGCAAGCAGUGA------------------------AUCGACUGCUUCGGCGAGCAGCAAGAAAACUGCUUUAGAGAUCGCACUCGAGCACCACCUAGCGGUGAUGCCAUUACUUACAG .((.(((.((((.------------------------.((((.(((.((...((((((........))))))....))..))).))))(((.((((....)))).))))))).))).)). ( -28.90) >consensus AUGCAAGCAGUGC________________________AUCAACUGCUUCGGCAAGCAGCAAGAGAACUGCUUUAGAGAUAGCACUCGAGCUCCACCUAGUGGUGAAGCCAUUAUUUACCG .((((((((((..............................)))))))....((((((........))))))........))).....(((.((((....)))).)))............ (-22.95 = -23.11 + 0.16)



| Location | 17,234,259 – 17,234,379 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17234259 120 + 27905053 UAUCUCUAAAGCAGUUCUCUUGCUGCUUGCCGAAGCAGUUGAUGCGCUGCUUGUCGAAGCAGUUGAUGCACUGCUUGCACAUUGCGCUCGCACAGUCAUCAACUGACUACCGGGUAUAUC ........(((((((......))))))).(((.(((((((((((.((((..((.(((.(((((.(.((((.....))))))))))..)))))))))))))))))).))..)))....... ( -42.50) >DroSec_CAF1 29923 96 + 1 UAUCUCUGAAGCAGUUCUGUGGCUGCUUGCCGAAGCAGUUGAU------------------------GAACUGCUUGCAUAUUGACCUCCUACAGACAUCAACUGACUACCGGGUAUAUC (((..((((((((((((...((((((((....))))))))...------------------------))))))))).....((((..((.....))..))))........)))..))).. ( -27.40) >DroSim_CAF1 31095 96 + 1 UAUCUCUGAAGCAGUUCUCUGGCUGCUUGCCGAAGCAGUUGAU------------------------GAACUGCUUGCAUAGUGUCCUCGCAUAGACAUCAACUGACUACCGGGUAUAUC (((..((((((((((((..(((((((((....)))))))))..------------------------))))))))).....(((((........)))))...........)))..))).. ( -30.20) >consensus UAUCUCUGAAGCAGUUCUCUGGCUGCUUGCCGAAGCAGUUGAU________________________GAACUGCUUGCAUAUUGACCUCGCACAGACAUCAACUGACUACCGGGUAUAUC ........(((((((......))))))).(((.((((((((((............................((....))...(((....))).....)))))))).))..)))....... (-21.26 = -21.27 + 0.00)



| Location | 17,234,259 – 17,234,379 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -19.57 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17234259 120 - 27905053 GAUAUACCCGGUAGUCAGUUGAUGACUGUGCGAGCGCAAUGUGCAAGCAGUGCAUCAACUGCUUCGACAAGCAGCGCAUCAACUGCUUCGGCAAGCAGCAAGAGAACUGCUUUAGAGAUA .......((((.((.(((((((((.((((.(((((((...)))).((((((......)))))))))....))))..))))))))))))))).((((((........))))))........ ( -43.40) >DroSec_CAF1 29923 96 - 1 GAUAUACCCGGUAGUCAGUUGAUGUCUGUAGGAGGUCAAUAUGCAAGCAGUUC------------------------AUCAACUGCUUCGGCAAGCAGCCACAGAACUGCUUCAGAGAUA .............(((.((((((.((.....)).)))))).((.(((((((((------------------------.....((((((....)))))).....)))))))))))..))). ( -29.30) >DroSim_CAF1 31095 96 - 1 GAUAUACCCGGUAGUCAGUUGAUGUCUAUGCGAGGACACUAUGCAAGCAGUUC------------------------AUCAACUGCUUCGGCAAGCAGCCAGAGAACUGCUUCAGAGAUA ......((((((((.((.....)).)))).)).))...((.((.(((((((((------------------------.((..((((((....))))))...))))))))))))).))... ( -29.90) >consensus GAUAUACCCGGUAGUCAGUUGAUGUCUGUGCGAGGACAAUAUGCAAGCAGUUC________________________AUCAACUGCUUCGGCAAGCAGCCAGAGAACUGCUUCAGAGAUA .......((((.((.((((((((..((.....)).......((....))............................)))))))))))))).((((((........))))))........ (-19.57 = -19.57 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:41 2006