
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,230,179 – 17,230,339 |
| Length | 160 |
| Max. P | 0.936954 |

| Location | 17,230,179 – 17,230,299 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -28.72 |
| Energy contribution | -29.52 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17230179 120 - 27905053 CUUCAGAGCCUCGCAGUCCUUCAAGAAGGCUGCGAGCAAUUUUAGAAUAUAGGACGACGGGAGCACCUGGCAGUGAUACAGAUCCACCAUGCAGUGAACCACUCCACGGCACCUGUUCCA .......((.((((((.((((....))))))))))))..............(((..(((((.((.(.(((.((((.(((..............)))...))))))).))).)))))))). ( -36.54) >DroSec_CAF1 25627 120 - 1 CUUCAGAGCCUCGCAGUCCUUCAUGAAGGCUGCGAGCAAAUUCAGAAUAUAGGACGACGAGAUCACCCGGCAGUUAUACAGGUCCACCAGGCAGUGGACCACUCCACGGCACUUGUUCCA .......((.((((((.((((....))))))))))))..............(((..(((((.....((((.(((......(((((((......)))))))))).).)))..)))))))). ( -39.20) >DroSim_CAF1 26810 120 - 1 CUUCAGAGCCUCGCAGUCCUUCAUGAAGGCUGCGAGCAAAUUCAGAAUAUAGGACGACGGGAUCACCCGGCAGUUAUACAGGUCCACCAGGCAGUGGACCACUCCACGGCACUUGUUCCA .......(((((((((.((((....))))))))))................(((...((((....))))...........(((((((......)))))))..)))..))).......... ( -42.20) >DroEre_CAF1 27429 120 - 1 CAUCAGAGCCUCGCACUCCUUUAGGAGGACUGCGAACAUUGUCAGCAUGGAGGACGACGGUAUCACUUGGCAGUUAUACAGGUCCACCAGGAAGUGGACCACACCUCGGCACCUGUCCCA ...(((.((((((((.(((((...))))).)))))..............((((......((((.(((....))).)))).(((((((......)))))))...)))))))..)))..... ( -41.10) >DroYak_CAF1 34356 120 - 1 CCUCAGAGCCUCGCACUCCUUUAGGAAGACUGCGAGGAGGUUCAGCAGGGAGGACGACGGAAUCACUUGGCAGUUGUACAGGUCCACCAGGAAGUGGACUACUCCCCGGCACUUGGACCA ........(((((((.(((....)))....))))))).((((((((.(((((.(((((..............)))))...(((((((......))))))).)))))..))...)))))). ( -46.24) >consensus CUUCAGAGCCUCGCAGUCCUUCAGGAAGGCUGCGAGCAAAUUCAGAAUAUAGGACGACGGGAUCACCUGGCAGUUAUACAGGUCCACCAGGCAGUGGACCACUCCACGGCACUUGUUCCA .......(((((((((((.........))))))))................(((...((((....))))...........(((((((......)))))))..)))..))).......... (-28.72 = -29.52 + 0.80)



| Location | 17,230,219 – 17,230,339 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -48.14 |
| Consensus MFE | -33.54 |
| Energy contribution | -34.22 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17230219 120 + 27905053 UGUAUCACUGCCAGGUGCUCCCGUCGUCCUAUAUUCUAAAAUUGCUCGCAGCCUUCUUGAAGGACUGCGAGGCUCUGAAGGAUCCGGACGAUCUGGUGGGCGUCACACCGCAGAUACGAC .(((((.(..((((........(((((((...(((((......((((((((((((....)))).)))))).)).....)))))..)))))))))))..)(((......))).)))))... ( -47.20) >DroSec_CAF1 25667 120 + 1 UGUAUAACUGCCGGGUGAUCUCGUCGUCCUAUAUUCUGAAUUUGCUCGCAGCCUUCAUGAAGGACUGCGAGGCUCUGAAGGAUCCGGACGAUCUGGUGGGCAUCACACCGCAGGUACGAC .((((..((((.(((((((((((((((((...(((((......((((((((((((....)))).)))))).)).....)))))..))))))).....))).))))).))))))))))... ( -48.50) >DroSim_CAF1 26850 120 + 1 UGUAUAACUGCCGGGUGAUCCCGUCGUCCUAUAUUCUGAAUUUGCUCGCAGCCUUCAUGAAGGACUGCGAGGCUCUGAAGGAUCCGGACGAUCUGGUGGGCAUCACACCGCAGGUACGAC .((((..((((.(((((((((((((((((...(((((......((((((((((((....)))).)))))).)).....)))))..))))))).....))).))))).))))))))))... ( -51.50) >DroEre_CAF1 27469 117 + 1 UGUAUAACUGCCAAGUGAUACCGUCGUCCUCCAUGCUGACAAUGUUCGCAGUCCUCCUAAAGGAGUGCGAGGCUCUGAUGGAUCCGGACGAUGUGGUGGUC---GCUCCGCAGCUGCGUC .(((...((((..((((((.((((((((((((((.(.((.....((((((.((((.....)))).))))))..)).))))))...)))))))).)...)))---)))..)))).)))... ( -45.60) >DroYak_CAF1 34396 120 + 1 UGUACAACUGCCAAGUGAUUCCGUCGUCCUCCCUGCUGAACCUCCUCGCAGUCUUCCUAAAGGAGUGCGAGGCUCUGAGGGAUCCGGACGAUGUGGUGGUAGCGGCUCCGCAGACACGUC (((...((..(((........(((((((((((((.(.((....(((((((.((((.....)))).))))))).)).))))))...)))))))))))..)).(((....)))..))).... ( -47.90) >consensus UGUAUAACUGCCAGGUGAUCCCGUCGUCCUAUAUUCUGAAAUUGCUCGCAGCCUUCAUGAAGGACUGCGAGGCUCUGAAGGAUCCGGACGAUCUGGUGGGCAUCACACCGCAGAUACGAC .((((..((((...(((..((((((((((................((((((((((....)))).))))))((.((.....)).))))))))).....)))...)))...))))))))... (-33.54 = -34.22 + 0.68)

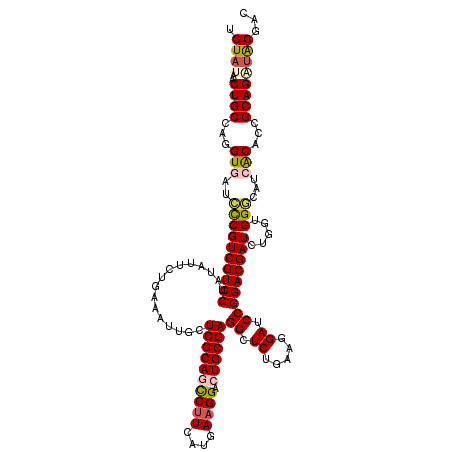
| Location | 17,230,219 – 17,230,339 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -42.93 |
| Consensus MFE | -31.56 |
| Energy contribution | -32.36 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17230219 120 - 27905053 GUCGUAUCUGCGGUGUGACGCCCACCAGAUCGUCCGGAUCCUUCAGAGCCUCGCAGUCCUUCAAGAAGGCUGCGAGCAAUUUUAGAAUAUAGGACGACGGGAGCACCUGGCAGUGAUACA ...(((((((((((((....(((....(.((((((..((.((.......(((((((.((((....))))))))))).......)).))...)))))))))).)))))..)))..))))). ( -45.24) >DroSec_CAF1 25667 120 - 1 GUCGUACCUGCGGUGUGAUGCCCACCAGAUCGUCCGGAUCCUUCAGAGCCUCGCAGUCCUUCAUGAAGGCUGCGAGCAAAUUCAGAAUAUAGGACGACGAGAUCACCCGGCAGUUAUACA ((.(((.((((((.(((((.(......(.((((((.((....))...((.((((((.((((....))))))))))))..............)))))))..)))))))).)))).))))). ( -40.40) >DroSim_CAF1 26850 120 - 1 GUCGUACCUGCGGUGUGAUGCCCACCAGAUCGUCCGGAUCCUUCAGAGCCUCGCAGUCCUUCAUGAAGGCUGCGAGCAAAUUCAGAAUAUAGGACGACGGGAUCACCCGGCAGUUAUACA ((.(((.((((((.(((((.(((....(.((((((.((....))...((.((((((.((((....))))))))))))..............))))))))))))))))).)))).))))). ( -46.60) >DroEre_CAF1 27469 117 - 1 GACGCAGCUGCGGAGC---GACCACCACAUCGUCCGGAUCCAUCAGAGCCUCGCACUCCUUUAGGAGGACUGCGAACAUUGUCAGCAUGGAGGACGACGGUAUCACUUGGCAGUUAUACA .....((((((.(((.---((..(((...((((((...((((((.((...(((((.(((((...))))).)))))...))....).))))))))))).))).)).))).))))))..... ( -41.40) >DroYak_CAF1 34396 120 - 1 GACGUGUCUGCGGAGCCGCUACCACCACAUCGUCCGGAUCCCUCAGAGCCUCGCACUCCUUUAGGAAGACUGCGAGGAGGUUCAGCAGGGAGGACGACGGAAUCACUUGGCAGUUGUACA ...(((.((((.((((((...........((((((...((((((.((((((((((.(((....)))....)))...))))))).).)))))))))))))).....))).))))...))). ( -41.00) >consensus GUCGUACCUGCGGUGUGACGCCCACCAGAUCGUCCGGAUCCUUCAGAGCCUCGCAGUCCUUCAGGAAGGCUGCGAGCAAAUUCAGAAUAUAGGACGACGGGAUCACCUGGCAGUUAUACA ...(((.((((((((.......))))...((((((((.((.....)).))((((((((.........))))))))................))))))((((....))))))))...))). (-31.56 = -32.36 + 0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:35 2006