
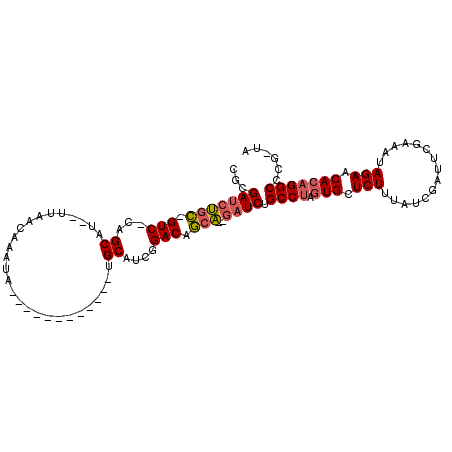
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,200,439 – 17,200,530 |
| Length | 91 |
| Max. P | 0.988065 |

| Location | 17,200,439 – 17,200,530 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -14.44 |
| Energy contribution | -14.72 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17200439 91 + 27905053 CGCGAUCUCC-GUC-CAGCAU--UUAACAAAUA------------UGCAUCGGACAGCA--GAUCUGCCUAGUGCUCUUUGUCGAUUCGAAAUAGAACACAGGCCCG-UA .((((((((.-(((-(.((((--.........)------------)))...)))).).)--)))).((((.(((.(((...(((...)))...))).)))))))..)-). ( -27.40) >DroSec_CAF1 22075 91 + 1 CGUGAUCUGC-GUC-CAGCAU--UUAACAAAUA------------UGCAGCGGACAGCA--GAUCUGCCUAGUGCUCUUUGUCGCUUCGAAAUAGAACACAGGCCCG-UA ...(((((((-(((-(.((((--.........)------------)))...)))).)))--)))).((((.(((.(((...(((...)))...))).)))))))...-.. ( -32.60) >DroSim_CAF1 22954 91 + 1 CGUGAUCUGC-GUC-CAGCAU--UUAACAAAUA------------UGCAUCGGACAGCA--GAUCUGCCUAGUGCUCUUUGUCGCUUCGAAAUAGAACACAGGCCCG-UA ...(((((((-(((-(.((((--.........)------------)))...)))).)))--)))).((((.(((.(((...(((...)))...))).)))))))...-.. ( -32.60) >DroEre_CAF1 24127 88 + 1 CGCGAUCUGC-GUC-CAGCAU--UUAGUAAAUA------------UGCA---GACAGCA--GAUCUGCCUAGUGCUCUUUAUCGAUUUCAAAUAGAACACAGGCUUG-UU ...(((((((-(((-..((((--.........)------------))).---))).)))--)))).((((.(((.(((...............))).)))))))...-.. ( -26.96) >DroYak_CAF1 23813 93 + 1 CGCGAUCUGC-GUC-CAGCAU--UUAGCAAAUA------------UGCAUCGGACAGCAAAAAUCUGCCUAGUGCUCUUUAUCGAUUUCAAAUAGAACACAGGCUGG-UA .......(((-(((-(.((((--.........)------------)))...)))).)))...(((.((((.(((.(((...............))).))))))).))-). ( -22.36) >DroAna_CAF1 25117 107 + 1 CGAGACCGGUUGUCUUAGCUUUCUUAACUAAUAUUUUGAUUUAAAUGCUUGAGACAACC--GAUCGGCCAGAUGUUCUUUAGCG-UUUAAAAUAGAACACAGGCUGGCUU ...((.(((((((((((((...........................)).))))))))))--).))((((((.((((((......-........))))))....)))))). ( -32.87) >consensus CGCGAUCUGC_GUC_CAGCAU__UUAACAAAUA____________UGCAUCGGACAGCA__GAUCUGCCUAGUGCUCUUUAUCGAUUCGAAAUAGAACACAGGCCCG_UA ...(((((((.(((...((...........................))....))).)))..)))).((((.(((.(((...............))).)))))))...... (-14.44 = -14.72 + 0.28)


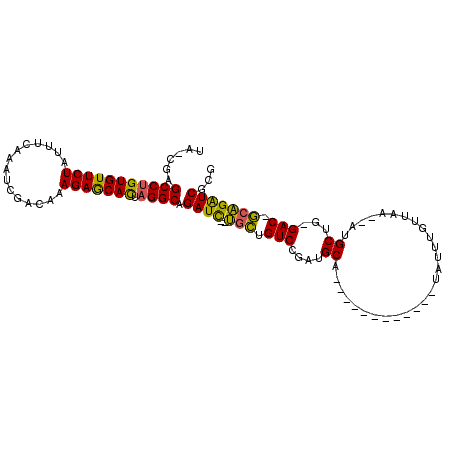
| Location | 17,200,439 – 17,200,530 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -35.01 |
| Consensus MFE | -21.85 |
| Energy contribution | -21.07 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.26 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17200439 91 - 27905053 UA-CGGGCCUGUGUUCUAUUUCGAAUCGACAAAGAGCACUAGGCAGAUC--UGCUGUCCGAUGCA------------UAUUUGUUAA--AUGCUG-GAC-GGAGAUCGCG ..-...(((((((((((.(.(((...))).).))))))).)))).((((--(.(((((((..(((------------(.........--))))))-)))-)))))))... ( -35.10) >DroSec_CAF1 22075 91 - 1 UA-CGGGCCUGUGUUCUAUUUCGAAGCGACAAAGAGCACUAGGCAGAUC--UGCUGUCCGCUGCA------------UAUUUGUUAA--AUGCUG-GAC-GCAGAUCACG ..-((.(((((((((((.(.(((...))).).))))))).)))).((((--(((.(((((..(((------------(.........--))))))-)))-))))))).)) ( -37.20) >DroSim_CAF1 22954 91 - 1 UA-CGGGCCUGUGUUCUAUUUCGAAGCGACAAAGAGCACUAGGCAGAUC--UGCUGUCCGAUGCA------------UAUUUGUUAA--AUGCUG-GAC-GCAGAUCACG ..-((.(((((((((((.(.(((...))).).))))))).)))).((((--(((.(((((..(((------------(.........--))))))-)))-))))))).)) ( -37.20) >DroEre_CAF1 24127 88 - 1 AA-CAAGCCUGUGUUCUAUUUGAAAUCGAUAAAGAGCACUAGGCAGAUC--UGCUGUC---UGCA------------UAUUUACUAA--AUGCUG-GAC-GCAGAUCGCG ..-...(((((((((((..(((....)))...))))))).)))).((((--(((.(((---((((------------(.........--)))).)-)))-)))))))... ( -36.10) >DroYak_CAF1 23813 93 - 1 UA-CCAGCCUGUGUUCUAUUUGAAAUCGAUAAAGAGCACUAGGCAGAUUUUUGCUGUCCGAUGCA------------UAUUUGCUAA--AUGCUG-GAC-GCAGAUCGCG ..-...(((((((((((..(((....)))...))))))).)))).((((..(((.(((((..(((------------(.........--))))))-)))-)))))))... ( -30.80) >DroAna_CAF1 25117 107 - 1 AAGCCAGCCUGUGUUCUAUUUUAAA-CGCUAAAGAACAUCUGGCCGAUC--GGUUGUCUCAAGCAUUUAAAUCAAAAUAUUAGUUAAGAAAGCUAAGACAACCGGUCUCG ..(((((...(((((((........-......)))))))))))).((((--((((((((..(((.(((...................))).))).))))))))))))... ( -33.65) >consensus UA_CGAGCCUGUGUUCUAUUUCAAAUCGACAAAGAGCACUAGGCAGAUC__UGCUGUCCGAUGCA____________UAUUUGUUAA__AUGCUG_GAC_GCAGAUCGCG ......(((((((((((...............))))))).)))).((((..(((.(((....((...........................))...))).)))))))... (-21.85 = -21.07 + -0.77)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:28 2006