
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,199,253 – 17,199,394 |
| Length | 141 |
| Max. P | 0.987284 |

| Location | 17,199,253 – 17,199,355 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.30 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -16.03 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
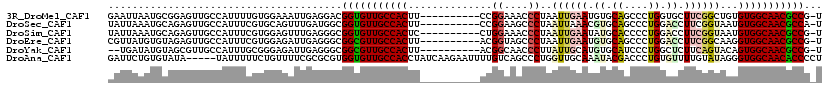
>3R_DroMel_CAF1 17199253 102 + 27905053 ACAAAUUGGCUUGCAUUUUUUUAAUGAAUUUGCA-CGGCGUUGCCACACAGCCGAAGCACCAGGGCUGCACAUUCAAUUAGGGUUUCCGG----------AAGUGGCAACACC ........(((((((..(((.....)))..))))-.)))((((((((.(((((..........)))))....(((.....((....)).)----------))))))))))... ( -31.80) >DroSec_CAF1 20769 102 + 1 ACAAAUUGGCUUGCAUUUUUUUUAUGAAUCUGCA-UGGCGUUGCCACAUUACCGAAGGUCCAGGGCUGCACGUUUAAUUAGGGCUUCCGG----------AAGUGGCAACACC ........(((((((..(((.....)))..))))-.)))((((((((.((.(((.((((((.((((.....)))).....)))))).)))----------))))))))))... ( -32.80) >DroSim_CAF1 21623 101 + 1 ACAAAUUGGCUUGCAUUUU-UUUAUGAAUCUGCA-CGGCGUUGCCACAUUACCGAAGGUCCAGGGGUGCAUAUUCAAUUAGGGUUUCCAG----------GAGUGGCAACACC ........(((((((..((-(....)))..))))-.)))((((((((.....(....)(((..(((.((.((......))..)).))).)----------))))))))))... ( -28.50) >DroEre_CAF1 22998 101 + 1 ACAAAUUGGCUUGCAUUAU-UUUAUGAUUUUACA-CGGCGUUGCCACCUUGCCGAAGGUCCAGGGCUGCACAUUCAAUUAGGGCUACCGU----------AAGUGGCAACGCC ...................-..............-.(((((((((((.((((.(((.((.((....)).)).))).....((....))))----------))))))))))))) ( -29.80) >DroYak_CAF1 22783 101 + 1 ACAAAUUUGCCUGCAUUAU-UUUAUGAUUUUGCA-CGGCGUUGCCACUGUACUGAAGAGCCAGGGAUGCACAUGCAAUAAGGGUUGCCGU----------AAGUGGCAACGCC ...........((((..((-(....)))..))))-.((((((((((((.(((.(...((((.....(((....))).....)))).).))----------))))))))))))) ( -36.10) >DroAna_CAF1 24265 109 + 1 ACAAAUG---CAGCAUAGG-UUUAUCAAUUUUUAGGGGUGUUGCCACCCUAUACAAAACACAGGGUCGUAUUUGCAACCAGGGCUGACAAAAUUCUUGAUAGGUGGCAACACC .......---.........-................((((((((((((.(((.(((........((((..((((....))))..)))).......)))))))))))))))))) ( -28.46) >consensus ACAAAUUGGCUUGCAUUUU_UUUAUGAAUUUGCA_CGGCGUUGCCACAUUACCGAAGGUCCAGGGCUGCACAUUCAAUUAGGGCUUCCGG__________AAGUGGCAACACC ....................................((.((((((((....((.........))................((....))..............)))))))).)) (-16.03 = -16.12 + 0.09)

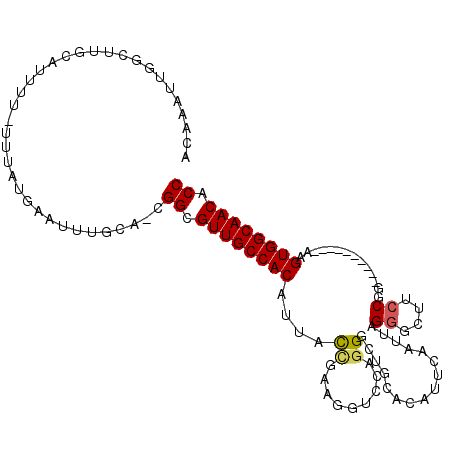
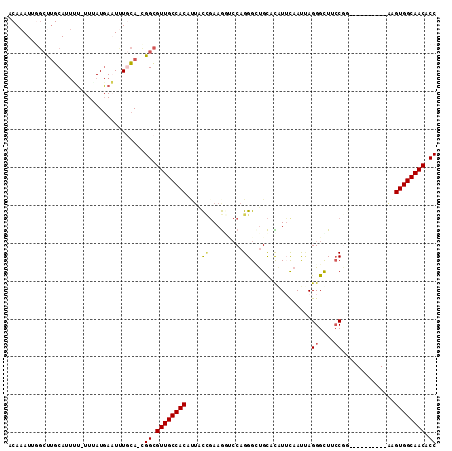
| Location | 17,199,253 – 17,199,355 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.30 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.75 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17199253 102 - 27905053 GGUGUUGCCACUU----------CCGGAAACCCUAAUUGAAUGUGCAGCCCUGGUGCUUCGGCUGUGUGGCAACGCCG-UGCAAAUUCAUUAAAAAAAUGCAAGCCAAUUUGU (((((((((((..----------..(....).............((((((..........))))))))))))))))).-.(((((((((((.....))))......))))))) ( -33.00) >DroSec_CAF1 20769 102 - 1 GGUGUUGCCACUU----------CCGGAAGCCCUAAUUAAACGUGCAGCCCUGGACCUUCGGUAAUGUGGCAACGCCA-UGCAGAUUCAUAAAAAAAAUGCAAGCCAAUUUGU (((((((((((..----------(((((.(.((...................)).).)))))....))))))))))).-((((...............))))........... ( -30.97) >DroSim_CAF1 21623 101 - 1 GGUGUUGCCACUC----------CUGGAAACCCUAAUUGAAUAUGCACCCCUGGACCUUCGGUAAUGUGGCAACGCCG-UGCAGAUUCAUAAA-AAAAUGCAAGCCAAUUUGU (((((((((((..----------..(....)....((((((..(.((....)).)..))))))...))))))))))).-((((..........-....))))........... ( -27.24) >DroEre_CAF1 22998 101 - 1 GGCGUUGCCACUU----------ACGGUAGCCCUAAUUGAAUGUGCAGCCCUGGACCUUCGGCAAGGUGGCAACGCCG-UGUAAAAUCAUAAA-AUAAUGCAAGCCAAUUUGU (((((((((((((----------..((....))...(((((.((.((....)).)).)))))..)))))))))))))(-((......)))...-.....(((((....))))) ( -31.50) >DroYak_CAF1 22783 101 - 1 GGCGUUGCCACUU----------ACGGCAACCCUUAUUGCAUGUGCAUCCCUGGCUCUUCAGUACAGUGGCAACGCCG-UGCAAAAUCAUAAA-AUAAUGCAGGCAAAUUUGU (((((((((((((----------(((....)((....(((....))).....)).......))).)))))))))))).-(((.....(((...-...)))...)))....... ( -33.00) >DroAna_CAF1 24265 109 - 1 GGUGUUGCCACCUAUCAAGAAUUUUGUCAGCCCUGGUUGCAAAUACGACCCUGUGUUUUGUAUAGGGUGGCAACACCCCUAAAAAUUGAUAAA-CCUAUGCUG---CAUUUGU ((((((((((((((((((((..((((.((((....))))))))((((....))))))))).))).))))))))))))................-.........---....... ( -34.20) >consensus GGUGUUGCCACUU__________CCGGAAACCCUAAUUGAAUGUGCAGCCCUGGACCUUCGGUAAUGUGGCAACGCCG_UGCAAAUUCAUAAA_AAAAUGCAAGCCAAUUUGU (((((((((((..............((....))..((((((.((.((....)).)).))))))...))))))))))).................................... (-21.86 = -22.75 + 0.89)

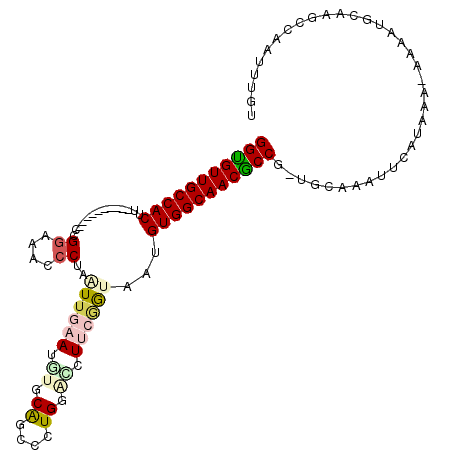
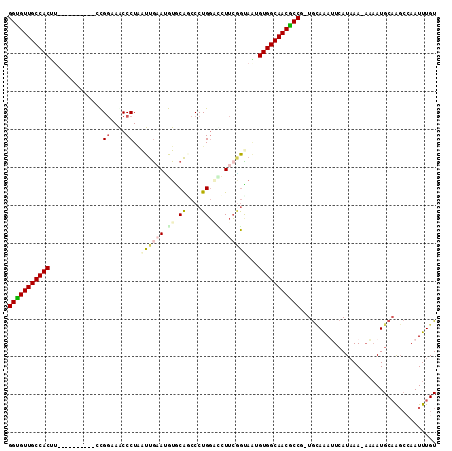
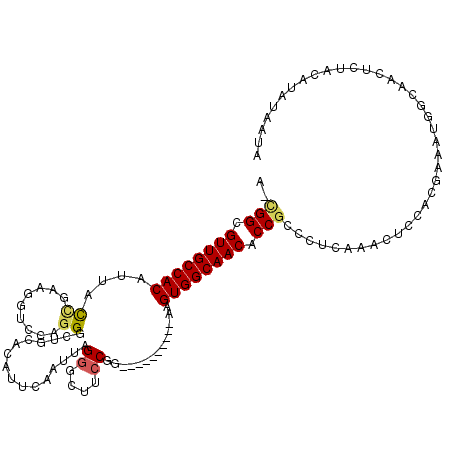
| Location | 17,199,286 – 17,199,394 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.87 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -17.83 |
| Energy contribution | -17.97 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17199286 108 + 27905053 A-CGGCGUUGCCACACAGCCGAAGCACCAGGGCUGCACAUUCAAUUAGGGUUUCCGG----------AAGUGGCAACACCGUCCUCAAUUUCCACAAAAUGGCAACUCCGCAUUAAUUC .-(((.(((((((..(((((..........)))))............(((.....((----------(.(((....)))..)))......)))......))))))).)))......... ( -33.70) >DroSec_CAF1 20802 108 + 1 A-UGGCGUUGCCACAUUACCGAAGGUCCAGGGCUGCACGUUUAAUUAGGGCUUCCGG----------AAGUGGCAACACCGCCAUCAAACUGCACGAAAUGGCAACUCUGCAUUUAAUA .-.((.((((((((.((.(((.((((((.((((.....)))).....)))))).)))----------)))))))))).))(((((.............)))))................ ( -33.92) >DroSim_CAF1 21655 108 + 1 A-CGGCGUUGCCACAUUACCGAAGGUCCAGGGGUGCAUAUUCAAUUAGGGUUUCCAG----------GAGUGGCAACACCGCCCUCAAACUCCACGAAAUGGCAACUCUGCAUUUAAUA .-(((.(((((((....(((...)))...(((((.............((....))..----------(((.(((......))))))..)))))......))))))).)))......... ( -29.90) >DroEre_CAF1 23030 108 + 1 A-CGGCGUUGCCACCUUGCCGAAGGUCCAGGGCUGCACAUUCAAUUAGGGCUACCGU----------AAGUGGCAACGCCGCCCUCAAUCUCCACGAAAUGGCAACUCUACACAUAACG .-.(((((((((((.((((.(((.((.((....)).)).))).....((....))))----------)))))))))))))(((.....((.....))...)))................ ( -34.40) >DroYak_CAF1 22815 106 + 1 A-CGGCGUUGCCACUGUACUGAAGAGCCAGGGAUGCACAUGCAAUAAGGGUUGCCGU----------AAGUGGCAACGCCGCCCUCAAUCUCCCGCAAAUGGCAACGCUACAUAUCA-- .-.((((((((((.(((...((.((...((((.(((....)))....(((((((((.----------...))))))).)).))))...))))..)))..))))))))))........-- ( -41.50) >DroAna_CAF1 24294 114 + 1 AGGGGUGUUGCCACCCUAUACAAAACACAGGGUCGUAUUUGCAACCAGGGCUGACAAAAUUCUUGAUAGGUGGCAACACCACGCGCGAAAACAGAAAAAUA-----UAUACACAGAAUC ...((((((((((((.(((.(((........((((..((((....))))..)))).......)))))))))))))))))).....................-----............. ( -30.16) >consensus A_CGGCGUUGCCACAUUACCGAAGGUCCAGGGCUGCACAUUCAAUUAGGGCUUCCGG__________AAGUGGCAACACCGCCCUCAAACUCCACGAAAUGGCAACUCUACAUAUAAUA ..(((.((((((((....((.........))................((....))..............)))))))).)))...................................... (-17.83 = -17.97 + 0.14)

| Location | 17,199,286 – 17,199,394 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.87 |
| Mean single sequence MFE | -38.49 |
| Consensus MFE | -22.11 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17199286 108 - 27905053 GAAUUAAUGCGGAGUUGCCAUUUUGUGGAAAUUGAGGACGGUGUUGCCACUU----------CCGGAAACCCUAAUUGAAUGUGCAGCCCUGGUGCUUCGGCUGUGUGGCAACGCCG-U .((((..((((((((....))))))))..))))....(((((((((((((..----------..(....).............((((((..........))))))))))))))))))-) ( -37.50) >DroSec_CAF1 20802 108 - 1 UAUUAAAUGCAGAGUUGCCAUUUCGUGCAGUUUGAUGGCGGUGUUGCCACUU----------CCGGAAGCCCUAAUUAAACGUGCAGCCCUGGACCUUCGGUAAUGUGGCAACGCCA-U ((((((((((((((.......))).)))).)))))))..(((((((((((..----------(((((.(.((...................)).).)))))....))))))))))).-. ( -36.81) >DroSim_CAF1 21655 108 - 1 UAUUAAAUGCAGAGUUGCCAUUUCGUGGAGUUUGAGGGCGGUGUUGCCACUC----------CUGGAAACCCUAAUUGAAUAUGCACCCCUGGACCUUCGGUAAUGUGGCAACGCCG-U .........((((....((((...))))..))))...(((((((((((((..----------..(....)....((((((..(.((....)).)..))))))...))))))))))))-) ( -31.30) >DroEre_CAF1 23030 108 - 1 CGUUAUGUGUAGAGUUGCCAUUUCGUGGAGAUUGAGGGCGGCGUUGCCACUU----------ACGGUAGCCCUAAUUGAAUGUGCAGCCCUGGACCUUCGGCAAGGUGGCAACGCCG-U .............((((((((((.((((((.((.(((((((.((((((....----------..))))))))....((......))))))).)).)))).)).))))))))))....-. ( -41.10) >DroYak_CAF1 22815 106 - 1 --UGAUAUGUAGCGUUGCCAUUUGCGGGAGAUUGAGGGCGGCGUUGCCACUU----------ACGGCAACCCUUAUUGCAUGUGCAUCCCUGGCUCUUCAGUACAGUGGCAACGCCG-U --.........((((((((((((((.(((((.(.((((.((.((((((....----------..))))))))....(((....))).)))).).))))).))).)))))))))))..-. ( -47.90) >DroAna_CAF1 24294 114 - 1 GAUUCUGUGUAUA-----UAUUUUUCUGUUUUCGCGCGUGGUGUUGCCACCUAUCAAGAAUUUUGUCAGCCCUGGUUGCAAAUACGACCCUGUGUUUUGUAUAGGGUGGCAACACCCCU ......((((...-----.((......))....))))(.((((((((((((((((((((..((((.((((....))))))))((((....))))))))).))).)))))))))))).). ( -36.30) >consensus GAUUAAAUGCAGAGUUGCCAUUUCGUGGAGAUUGAGGGCGGUGUUGCCACUU__________CCGGAAACCCUAAUUGAAUGUGCAGCCCUGGACCUUCGGUAAUGUGGCAACGCCG_U .......................................(((((((((((..............((....))..((((((.((.((....)).)).))))))...)))))))))))... (-22.11 = -23.00 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:24 2006