
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,189,741 – 17,189,835 |
| Length | 94 |
| Max. P | 0.765013 |

| Location | 17,189,741 – 17,189,835 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -15.26 |
| Energy contribution | -18.04 |
| Covariance contribution | 2.78 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
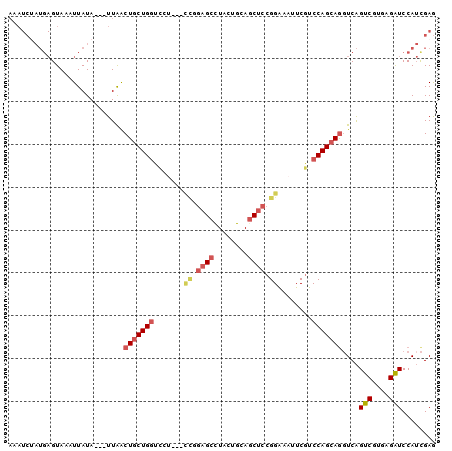
>3R_DroMel_CAF1 17189741 94 + 27905053 AAAUCGAUGAGUAUAUUAUA---UUAACUGCUGGUCCU---CCGGAGCUUACUGCAGCUCCGGAAAUUCGUCCAGCAGGUCAGUCGUGAGAUCCAUCGAG ...((((((...........---....(((((((.(.(---((((((((......))))))))).....).)))))))....(((....))).)))))). ( -40.10) >DroPse_CAF1 13262 90 + 1 ----------GUAAAUUAUAAGUUUGGCUACUGCUCAACGAUGGACGACUACUGCAACUCGGGAAAAUCAUCCAGCAGGUCGGCCGAAAGGUCCAGCGAA ----------...........(((.(((....))).)))(.(((.((((..((((......(((......))).))))))))(((....)))))).)... ( -24.10) >DroSec_CAF1 11210 94 + 1 AAAUCUAUGAGUAUAUUAUA---UUAACUGCUGGUCCU---CCGGAGCCUACUGCAGCUCCGGAAAUUCGUCCAGCAGGUCAGUCGUGAGAUCCAUCGAG ...((.(((...........---....(((((((.(.(---(((((((........)))))))).....).)))))))....(((....))).))).)). ( -32.80) >DroSim_CAF1 12080 94 + 1 AAAUCUAUGAGUAUAUUAUA---UUAACUGCUGGUCCU---CCGGAGCCUACUGCAGCUCCGGAAAUUCGUCCAGCAGGUCAGUCGUGAGAUCCAUCGAG ...((.(((...........---....(((((((.(.(---(((((((........)))))))).....).)))))))....(((....))).))).)). ( -32.80) >DroEre_CAF1 13230 94 + 1 AAAUCUAUGAGUAAAUUAUA---UUAACUGCUGGUCGU---AUAGAGCCUAUUGCAGCUCCGGAAAUUCGUCCAGCAGUUCAGUCGCGAGAUCCAUUGAG ...((.(((...........---..(((((((((.((.---...((((........))))........)).)))))))))..(((....))).))).)). ( -25.10) >DroYak_CAF1 13024 94 + 1 AAAUCUAUGAGUAAAUUAUA---UUAACUGCUGGUUGA---UCAGAGCCUACUGCAGCUCCGGGAAUUCGUCCAGCAAGUCAGUCGUGAGAUCCAUCGAG ...((.(((...........---.....((((((.(((---((.((((........))))...))..))).)))))).....(((....))).))).)). ( -21.60) >consensus AAAUCUAUGAGUAAAUUAUA___UUAACUGCUGGUCCU___CCGGAGCCUACUGCAGCUCCGGAAAUUCGUCCAGCAGGUCAGUCGUGAGAUCCAUCGAG ...........................(((((((.......((.((((........)))).))........)))))))....(((....)))........ (-15.26 = -18.04 + 2.78)


| Location | 17,189,741 – 17,189,835 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -27.89 |
| Consensus MFE | -18.35 |
| Energy contribution | -18.21 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17189741 94 - 27905053 CUCGAUGGAUCUCACGACUGACCUGCUGGACGAAUUUCCGGAGCUGCAGUAAGCUCCGG---AGGACCAGCAGUUAA---UAUAAUAUACUCAUCGAUUU .(((((((...(((....))).(((((((.....(((((((((((......))))))))---))).)))))))....---..........)))))))... ( -36.90) >DroPse_CAF1 13262 90 - 1 UUCGCUGGACCUUUCGGCCGACCUGCUGGAUGAUUUUCCCGAGUUGCAGUAGUCGUCCAUCGUUGAGCAGUAGCCAAACUUAUAAUUUAC---------- ...(((((.((....))))...(((((.((((((.....(((.(......).)))...)))))).))))).)))................---------- ( -21.30) >DroSec_CAF1 11210 94 - 1 CUCGAUGGAUCUCACGACUGACCUGCUGGACGAAUUUCCGGAGCUGCAGUAGGCUCCGG---AGGACCAGCAGUUAA---UAUAAUAUACUCAUAGAUUU .(((.((.....))))).((((.((((((.....(((((((((((......))))))))---))).)))))))))).---.................... ( -30.40) >DroSim_CAF1 12080 94 - 1 CUCGAUGGAUCUCACGACUGACCUGCUGGACGAAUUUCCGGAGCUGCAGUAGGCUCCGG---AGGACCAGCAGUUAA---UAUAAUAUACUCAUAGAUUU .(((.((.....))))).((((.((((((.....(((((((((((......))))))))---))).)))))))))).---.................... ( -30.40) >DroEre_CAF1 13230 94 - 1 CUCAAUGGAUCUCGCGACUGAACUGCUGGACGAAUUUCCGGAGCUGCAAUAGGCUCUAU---ACGACCAGCAGUUAA---UAUAAUUUACUCAUAGAUUU .((.((((............(((((((((.((.((....((((((......))))))))---.)).)))))))))..---..........)))).))... ( -25.15) >DroYak_CAF1 13024 94 - 1 CUCGAUGGAUCUCACGACUGACUUGCUGGACGAAUUCCCGGAGCUGCAGUAGGCUCUGA---UCAACCAGCAGUUAA---UAUAAUUUACUCAUAGAUUU .(((.((.....))))).(((((.(((((..((.....(((((((......))))))).---))..)))))))))).---.................... ( -23.20) >consensus CUCGAUGGAUCUCACGACUGACCUGCUGGACGAAUUUCCGGAGCUGCAGUAGGCUCCGG___AGGACCAGCAGUUAA___UAUAAUAUACUCAUAGAUUU ......((.((....))))...(((((((.........(((((((......)))))))........)))))))........................... (-18.35 = -18.21 + -0.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:19 2006