
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,165,271 – 17,165,391 |
| Length | 120 |
| Max. P | 0.996982 |

| Location | 17,165,271 – 17,165,364 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.45 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17165271 93 + 27905053 CAUGCGCAGGAUCAGCGC------AGCGCUCUGAACUCUGAGCCCCAACUGAGCCGCUGCUCAGACCUCCGACUAAAGUCGA----ACAUAUAUAUCUGGCUA ..(((((.......))))------)..((((........)))).(((.((((((....)))))).....((((....)))).----...........)))... ( -28.20) >DroPse_CAF1 73082 83 + 1 CAUGCGCAGGAUCAGCGCAGGAGCAGCGCUCUGAACUCUGAGCCCCGACUGAGCCGCAGCUCAGACCUCGA--------------------GACCUCUGGCCG ..(((((.......)))))((..(((.(.(((((..(((((((..((.......))..)))))))..)).)--------------------)).).))).)). ( -29.40) >DroSec_CAF1 74530 77 + 1 CAUGCGCAGGAUCAGCGC------AGCGCUCUGAACUCUGAGCCCCAACUGAGCCGCUGCUCAGACCUCCGACU--------------------AUCUGGCUA ...((((.......))))------(((((((........)))).....((((((....))))))..........--------------------.....))). ( -23.90) >DroEre_CAF1 65627 93 + 1 CAUGCGCAGGAUCAGCGC------AGCGCUCUGAACUCUGAGCCCCAACUGAGCCGCUGCUCAGACCUCCGACUAAAGUCGA----ACAUAUAUAUCUGGCCA ..(((((.......))))------)..((((........)))).(((.((((((....)))))).....((((....)))).----...........)))... ( -28.20) >DroYak_CAF1 93139 97 + 1 CAUGCGCAGGAUCAGCGC------AGCCCUCUGAACUCUGAGCCCCAACUGAGCCGCUGCUCAGAGCUCUGACUAAAGUCGAACGAACAUAUAUAUCUGGCUA ...((((.......))))------((((....((.((((((((..(.........)..)))))))).))((((....)))).................)))). ( -27.50) >DroPer_CAF1 84968 83 + 1 CAUGCGCAGGAUCAGCGCAGGAGCAGCGCUCUGAACUCUGAGCCCCGACUGAGCCGCAGCUCAGACCUCGA--------------------GACCUCUGGCCG ..(((((.......)))))((..(((.(.(((((..(((((((..((.......))..)))))))..)).)--------------------)).).))).)). ( -29.40) >consensus CAUGCGCAGGAUCAGCGC______AGCGCUCUGAACUCUGAGCCCCAACUGAGCCGCUGCUCAGACCUCCGACU_________________UAUAUCUGGCCA ...((((.......)))).......((((((........)))).....((((((....))))))...................................)).. (-20.28 = -20.45 + 0.17)

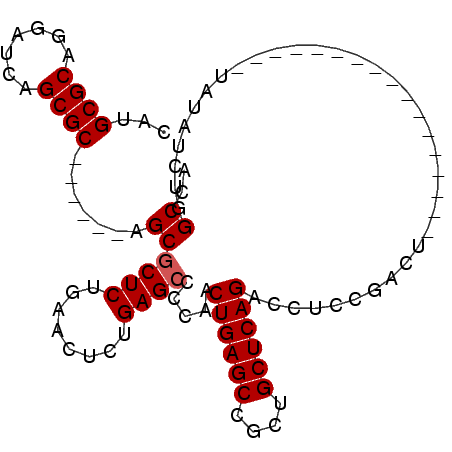
| Location | 17,165,271 – 17,165,364 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -36.15 |
| Consensus MFE | -31.04 |
| Energy contribution | -30.98 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17165271 93 - 27905053 UAGCCAGAUAUAUAUGU----UCGACUUUAGUCGGAGGUCUGAGCAGCGGCUCAGUUGGGGCUCAGAGUUCAGAGCGCU------GCGCUGAUCCUGCGCAUG ..(((((.........(----(((((....))))))((((...((((((.(((.....((((.....)))).)))))))------))...))))))).))... ( -33.50) >DroPse_CAF1 73082 83 - 1 CGGCCAGAGGUC--------------------UCGAGGUCUGAGCUGCGGCUCAGUCGGGGCUCAGAGUUCAGAGCGCUGCUCCUGCGCUGAUCCUGCGCAUG ..(((((.((((--------------------..(((.((((((((.((((...)))).)))))))).)))..(((((.......)))))))))))).))... ( -41.90) >DroSec_CAF1 74530 77 - 1 UAGCCAGAU--------------------AGUCGGAGGUCUGAGCAGCGGCUCAGUUGGGGCUCAGAGUUCAGAGCGCU------GCGCUGAUCCUGCGCAUG ..(((((..--------------------.(((.(((.(((((((..((((...))))..))))))).)))..((((..------.))))))).))).))... ( -27.60) >DroEre_CAF1 65627 93 - 1 UGGCCAGAUAUAUAUGU----UCGACUUUAGUCGGAGGUCUGAGCAGCGGCUCAGUUGGGGCUCAGAGUUCAGAGCGCU------GCGCUGAUCCUGCGCAUG ..(((((.........(----(((((....))))))((((...((((((.(((.....((((.....)))).)))))))------))...))))))).))... ( -33.30) >DroYak_CAF1 93139 97 - 1 UAGCCAGAUAUAUAUGUUCGUUCGACUUUAGUCAGAGCUCUGAGCAGCGGCUCAGUUGGGGCUCAGAGUUCAGAGGGCU------GCGCUGAUCCUGCGCAUG .((((..................(((....))).(((((((((((..((((...))))..)))))))))))....))))------((((.......))))... ( -38.70) >DroPer_CAF1 84968 83 - 1 CGGCCAGAGGUC--------------------UCGAGGUCUGAGCUGCGGCUCAGUCGGGGCUCAGAGUUCAGAGCGCUGCUCCUGCGCUGAUCCUGCGCAUG ..(((((.((((--------------------..(((.((((((((.((((...)))).)))))))).)))..(((((.......)))))))))))).))... ( -41.90) >consensus UAGCCAGAUAUA_________________AGUCGGAGGUCUGAGCAGCGGCUCAGUUGGGGCUCAGAGUUCAGAGCGCU______GCGCUGAUCCUGCGCAUG ..(((((...........................(((.(((((((..((((...))))..))))))).)))..(((((.......)))))....))).))... (-31.04 = -30.98 + -0.06)


| Location | 17,165,301 – 17,165,391 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -15.80 |
| Energy contribution | -17.68 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17165301 90 + 27905053 UCUGAGCCCCAACUGAGCCGCUGCUCAGACCUCCGACUAAAGUCGA----ACAUAUAUAUCUGGCUAUAGCCAG--UCAUACACUUACACAAUAUA ..((((......((((((....)))))).....((((....)))).----.....(((..(((((....)))))--..)))..))))......... ( -22.40) >DroSec_CAF1 74560 70 + 1 UCUGAGCCCCAACUGAGCCGCUGCUCAGACCUCCGACU--------------------AUCUGGCUAUAGCC------AUACACUUACACAAUAUA ..((((......((((((....))))))..........--------------------...((((....)))------)....))))......... ( -14.80) >DroEre_CAF1 65657 92 + 1 UCUGAGCCCCAACUGAGCCGCUGCUCAGACCUCCGACUAAAGUCGA----ACAUAUAUAUCUGGCCACAGCCAGAGCUAUACACUUACACAAUAUA ....(((.....((((((....)))))).....((((....)))).----.........((((((....))))))))).................. ( -24.80) >DroYak_CAF1 93169 96 + 1 UCUGAGCCCCAACUGAGCCGCUGCUCAGAGCUCUGACUAAAGUCGAACGAACAUAUAUAUCUGGCUAUAGCCAGAGCUAUACACUUACACAAUAUA ..((((......((((((....))))))(((((((.(((.(((((................))))).))).))))))).....))))......... ( -26.79) >consensus UCUGAGCCCCAACUGAGCCGCUGCUCAGACCUCCGACUAAAGUCGA____ACAUAUAUAUCUGGCUAUAGCCAG__CUAUACACUUACACAAUAUA ..((((......((((((....)))))).....((((....))))..............((((((....))))))........))))......... (-15.80 = -17.68 + 1.87)



| Location | 17,165,301 – 17,165,391 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -20.70 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17165301 90 - 27905053 UAUAUUGUGUAAGUGUAUGA--CUGGCUAUAGCCAGAUAUAUAUGU----UCGACUUUAGUCGGAGGUCUGAGCAGCGGCUCAGUUGGGGCUCAGA ............(((((((.--(((((....))))).))))))).(----(((((....))))))..(((((((..((((...))))..))))))) ( -32.90) >DroSec_CAF1 74560 70 - 1 UAUAUUGUGUAAGUGUAU------GGCUAUAGCCAGAU--------------------AGUCGGAGGUCUGAGCAGCGGCUCAGUUGGGGCUCAGA ((((((.....))))))(------((((((......))--------------------)))))....(((((((..((((...))))..))))))) ( -20.20) >DroEre_CAF1 65657 92 - 1 UAUAUUGUGUAAGUGUAUAGCUCUGGCUGUGGCCAGAUAUAUAUGU----UCGACUUUAGUCGGAGGUCUGAGCAGCGGCUCAGUUGGGGCUCAGA ............(((((((..((((((....))))))))))))).(----(((((....))))))..(((((((..((((...))))..))))))) ( -33.00) >DroYak_CAF1 93169 96 - 1 UAUAUUGUGUAAGUGUAUAGCUCUGGCUAUAGCCAGAUAUAUAUGUUCGUUCGACUUUAGUCAGAGCUCUGAGCAGCGGCUCAGUUGGGGCUCAGA ........(...(((((((..((((((....)))))))))))))...)(((((((....))).))))(((((((..((((...))))..))))))) ( -33.00) >consensus UAUAUUGUGUAAGUGUAUAG__CUGGCUAUAGCCAGAUAUAUAUGU____UCGACUUUAGUCGGAGGUCUGAGCAGCGGCUCAGUUGGGGCUCAGA ((((((.....)))))).....(((((....)))))...............................(((((((..((((...))))..))))))) (-20.70 = -21.20 + 0.50)

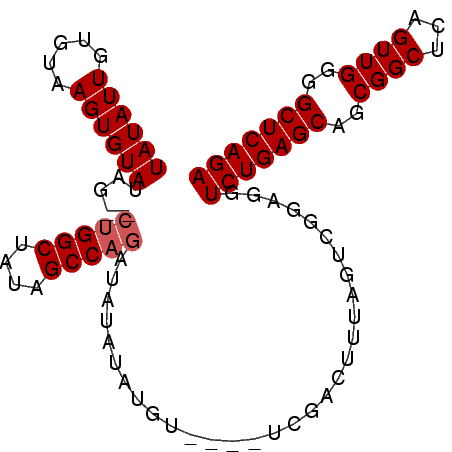

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:11 2006