
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,160,956 – 17,161,059 |
| Length | 103 |
| Max. P | 0.997100 |

| Location | 17,160,956 – 17,161,059 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.64 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -13.80 |
| Energy contribution | -15.47 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
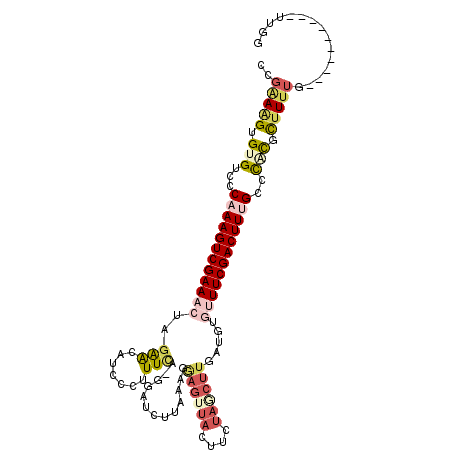
>3R_DroMel_CAF1 17160956 103 + 27905053 CCGAAAGUGUGUCCCAAAGUCGAAACAAGAACCUCCCUUUUAAGGAUCAUAAAGGAGUUACUUGUAACUUAAUGUGUUUCGACUUUGCUUACGCUUUUG----------UUGG .((((((((((...(((((((((((((.((.(((........))).))......((((((....))))))....)))))))))))))..))))))))))----------.... ( -37.20) >DroSec_CAF1 70167 106 + 1 CCGAAAGUGUGUCCCAAAGUCGAAACUAGAACAUCCCUUUCUAGGAUCUUAACGGAGUUACUUUUAGCUUGAUGUGUUUCGACUUUGCCCACGUUUUUUUUU-------UUUG ..(((((((((...((((((((((((....((((((((....))).........((((((....)))))))))))))))))))))))..)))))))))....-------.... ( -30.00) >DroSim_CAF1 61470 108 + 1 CCGGAAGUGUGUCCCAAAGUCGAAACUAGAACAUACCGUUCAAGGAUCUUAACGGAGUUACUUUUAGCUUGA-----UUCGACUUUGCCCACUUUUUUUUUUUUGUUUUUUGG ..(((((((.(...((((((((((.((.((((.....)))).))..........((((((....))))))..-----)))))))))).))))))))................. ( -25.30) >DroEre_CAF1 61364 102 + 1 CCGGAAGUGAGUCCCAAAGUCGAAACUAUAACAUCCCUUUCU-GUAGUUUAAAGGAGUUACUUCUAGCUUGAUGUGUUUCGACUUCGCUCACGCUUUUC----------UUGG ((((((((((((....((((((((((....((((((((((..-.......)))))(((((....))))).))))))))))))))).))))))....)))----------.))) ( -28.40) >DroYak_CAF1 88932 102 + 1 CCGGAAGUGAGUCCCAAAGUCGAAACUAGAACAGCCCUUUCU-GUAUUUUAAAGAAGUUACAUCUAUCUUGAUUUGUUUCGACUUCGCUCACUCCUUUG----------UUGG ..(((.((((((....((((((((((....((((......))-))......((((((......)).)))).....)))))))))).)))))))))....----------.... ( -28.30) >DroAna_CAF1 63370 87 + 1 CCAAAGGUGUGGCUCAAAGUCGAAGCUUGGGGAUUUCCUUUA-GCAGAU---------------UAUCCUUGUGAAGUUCGACUUUGUCCGUUCUUCAG----------UGAG .((.(((..(((..(((((((((((((..(((....)))..)-))...(---------------(((....))))..)))))))))).)))..)))...----------)).. ( -25.40) >consensus CCGAAAGUGUGUCCCAAAGUCGAAACUAGAACAUCCCUUUCA_GGAUCUUAAAGGAGUUACUUCUAGCUUGAUGUGUUUCGACUUUGCCCACGCUUUUG__________UUGG ..(((((.(((...((((((((((((..(((.......))).............((((((....)))))).....))))))))))))..))).)))))............... (-13.80 = -15.47 + 1.67)

| Location | 17,160,956 – 17,161,059 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.64 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -13.08 |
| Energy contribution | -12.89 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17160956 103 - 27905053 CCAA----------CAAAAGCGUAAGCAAAGUCGAAACACAUUAAGUUACAAGUAACUCCUUUAUGAUCCUUAAAAGGGAGGUUCUUGUUUCGACUUUGGGACACACUUUCGG ((..----------.....((....))((((((((((((.....(((((....))))).......((.((((......)))).)).))))))))))))))............. ( -27.90) >DroSec_CAF1 70167 106 - 1 CAAA-------AAAAAAAAACGUGGGCAAAGUCGAAACACAUCAAGCUAAAAGUAACUCCGUUAAGAUCCUAGAAAGGGAUGUUCUAGUUUCGACUUUGGGACACACUUUCGG ....-------..........(((..(((((((((((((((((..........((((...))))....(((....))))))))....))))))))))))...)))........ ( -26.20) >DroSim_CAF1 61470 108 - 1 CCAAAAAACAAAAAAAAAAAAGUGGGCAAAGUCGAA-----UCAAGCUAAAAGUAACUCCGUUAAGAUCCUUGAACGGUAUGUUCUAGUUUCGACUUUGGGACACACUUCCGG ...................((((((.((((((((((-----....(((...((.(((.((((((((...)))).))))...))))))))))))))))))...).))))).... ( -23.40) >DroEre_CAF1 61364 102 - 1 CCAA----------GAAAAGCGUGAGCGAAGUCGAAACACAUCAAGCUAGAAGUAACUCCUUUAAACUAC-AGAAAGGGAUGUUAUAGUUUCGACUUUGGGACUCACUUCCGG ....----------.......(((((((((((((((((...((......)).(((((((((((.......-...)))))).))))).))))))))))))...)))))...... ( -30.50) >DroYak_CAF1 88932 102 - 1 CCAA----------CAAAGGAGUGAGCGAAGUCGAAACAAAUCAAGAUAGAUGUAACUUCUUUAAAAUAC-AGAAAGGGCUGUUCUAGUUUCGACUUUGGGACUCACUUCCGG ....----------....((((((((((((((((((((.....((((.((......))))))......((-((......))))....))))))))))))...))))))))... ( -30.20) >DroAna_CAF1 63370 87 - 1 CUCA----------CUGAAGAACGGACAAAGUCGAACUUCACAAGGAUA---------------AUCUGC-UAAAGGAAAUCCCCAAGCUUCGACUUUGAGCCACACCUUUGG ....----------(..(((...((.(((((((((((((.....((((.---------------.(((..-...)))..))))..))).))))))))))..))....)))..) ( -22.70) >consensus CCAA__________CAAAAGAGUGAGCAAAGUCGAAACACAUCAAGCUAAAAGUAACUCCUUUAAGAUCC_AGAAAGGGAUGUUCUAGUUUCGACUUUGGGACACACUUCCGG ..........................((((((((((((.................................................))))))))))))(((......))).. (-13.08 = -12.89 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:02 2006