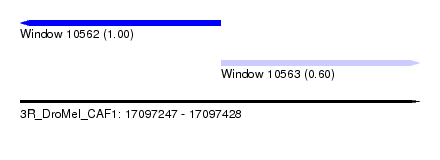
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,097,247 – 17,097,428 |
| Length | 181 |
| Max. P | 0.998210 |

| Location | 17,097,247 – 17,097,338 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.47 |
| Mean single sequence MFE | -43.90 |
| Consensus MFE | -26.30 |
| Energy contribution | -26.11 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.60 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
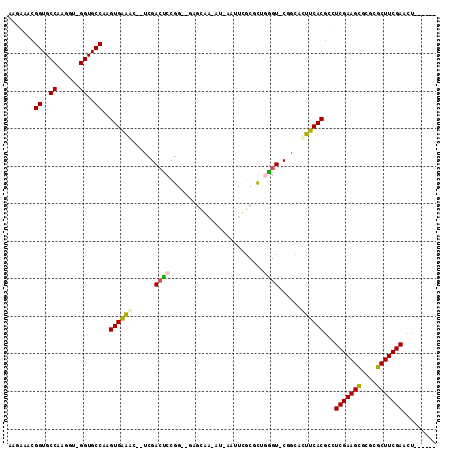
>3R_DroMel_CAF1 17097247 91 - 27905053 GAGAAACGGUGCCAAGGU-GGUGCCUCGUGAACG--UCGACUCCGG-------------AUUCGCCCGGGGU-CUGCACUUCACGCCUCGAAGCGCGCGCUUCGAACU------ .......((..((.....-))..)).((((((.(--(.((((((((-------------......)))))))-).))..))))))..(((((((....)))))))...------ ( -41.80) >DroPse_CAF1 9120 111 - 1 AGAAAACGGUGCCAAGGUAGGUGCCAAGUGAAACGCUAGACCCUUG-UGAGCAA-AUCAAUUUGCGAUGGGU-CGGCGUUUCACGCUUCGAAGCGCGCGCUUCGAACUACAGAU .......((..((......))..))..((((((((((.((((((((-..((...-.....))..))).))))-)))))))))))..((((((((....))))))))........ ( -49.90) >DroGri_CAF1 9416 104 - 1 CGGAAACGGUGCCAAGGU-GGUGCCAAGUGGAACG-GUGACGCCGGGAACGUGAAAUCAAUUCGCGCUUUGUUCACCAUUUCACACAUCGAAGUGCGCGCUUCGAG-------- .(....)((..((.....-))..))..((((((.(-((((.((..((..((((((.....))))))))..))))))).))))))...(((((((....))))))).-------- ( -41.80) >DroYak_CAF1 11796 89 - 1 UAGAAACGGUGCCAAGGU-GGUGCCUCGUGAACG--UCGACUCCGG-------------AUUCGCACGGGGU-CAGCACUUCACGCCUCGAAGCG--CGCUUCGAACU------ .......((..((.....-))..)).((((((.(--(.(((((((.-------------.......))))))-).))..))))))..((((((..--..))))))...------ ( -36.20) >DroAna_CAF1 9003 101 - 1 GAGAAACGGUGCCAAGGC-GGUGCCAAGUGAGGG--CCGAGUCCGG-GGAGCAA-AU-AAUUUGCACUCGGU-CGGAACUUCACGCCUCGAAGCGCGCGCUUCGAACU------ .......((..((.....-))..))..((((((.--((((..((((-(..((((-..-...)))).))))))-)))..))))))...(((((((....)))))))...------ ( -43.80) >DroPer_CAF1 9122 111 - 1 AGAAAACGGUGCCAAGGUAGGUGCCAAGUGAAACGCUAGACCCUUG-UGAGCAA-AUCAAUUUGCGAUGGGU-CGGCGUUUCACGCUUCGAAGCGCGCGCUUCGAACUACAGAU .......((..((......))..))..((((((((((.((((((((-..((...-.....))..))).))))-)))))))))))..((((((((....))))))))........ ( -49.90) >consensus AAGAAACGGUGCCAAGGU_GGUGCCAAGUGAAAC__UCGACUCCGG__GAGCAA_AU_AAUUCGCGCUGGGU_CGGCACUUCACGCCUCGAAGCGCGCGCUUCGAACU______ .......((..((......))..))..((((((......((((.........................))))......))))))...(((((((....)))))))......... (-26.30 = -26.11 + -0.19)


| Location | 17,097,338 – 17,097,428 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -13.82 |
| Energy contribution | -13.68 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
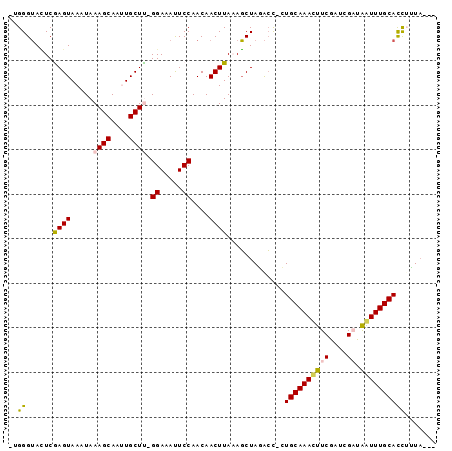
>3R_DroMel_CAF1 17097338 90 + 27905053 -UGGGUACUCGAGUAAAUAAAGCAAUUGCUC-GGAAAUUCCAACAACUUAAAGCUAGACC-CUGCAAACUUCGAUCGAUGGUUUGCACCUUUA--- -.((((....((((((.........))))))-((.....))................)))-)((((((((((....)).))))))))......--- ( -22.00) >DroPse_CAF1 9231 95 + 1 -CGGGUAUUCAAGUAAAUAAAGCAUCUGCUUAGGAAAUUCCAACAACUUAAAGCUAUACUUGUGCAAAUUUCGAUCGUCAAUUUGCACCAGUAUGA -..(((.((.((((.....((((....)))).((.....))....)))))).)))(((((.(((((((((.(....)..))))))))).))))).. ( -21.30) >DroSec_CAF1 9574 90 + 1 -AGGGUACUCGAGUAAAUAAAGCAAUUGCUC-GGAAACUCCAACAACUUAAAGCUAGACC-CUGCAAACUUCGAUCGAUAAUUUGCACCUUUA--- -(((((....((((((.........))))))-((.....))................)))-))(((((..((....))...))))).......--- ( -18.50) >DroEre_CAF1 9779 90 + 1 -UGGGUACUCGAGUAAAUAAAGCAAUUGCUU-GGAAAUUCCAACAACUUACAGCUAGACC-CUGCAAACUUCGAUCGAUAGUUUGCACCUUUA--- -.((((......((((...((((....))))-((.....))......))))......)))-)((((((((((....)).))))))))......--- ( -22.20) >DroAna_CAF1 9104 95 + 1 GUGGGAAAUCGAGUAAAUAAAGCACUUGCUUAGGAAAUUCCAACAACUUACAUCUAAGGC-AUGCAAAUUUCGAUCGAUAAUUUGCAUUUUUUUUU (((((..............((((....)))).((.....)).....)))))....((((.-(((((((((((....)).))))))))).))))... ( -17.80) >DroPer_CAF1 9233 95 + 1 -CGGGUAUUCAAGUAAAUAAAGCAUCUGCUUAGGAAAUUCCAACAACUUAAAGCUAUACUUGUGCAAAUUUCGAUCGUCAAUUUGCACCAGUAUGA -..(((.((.((((.....((((....)))).((.....))....)))))).)))(((((.(((((((((.(....)..))))))))).))))).. ( -21.30) >consensus _UGGGUACUCGAGUAAAUAAAGCAAUUGCUU_GGAAAUUCCAACAACUUAAAGCUAGACC_CUGCAAACUUCGAUCGAUAAUUUGCACCUUUA___ ..((......((((.....((((....)))).((.....))....)))).............((((((((((....)).))))))))))....... (-13.82 = -13.68 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:41 2006