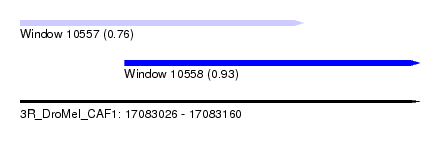
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,083,026 – 17,083,160 |
| Length | 134 |
| Max. P | 0.926808 |

| Location | 17,083,026 – 17,083,121 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -20.29 |
| Consensus MFE | -12.31 |
| Energy contribution | -13.07 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
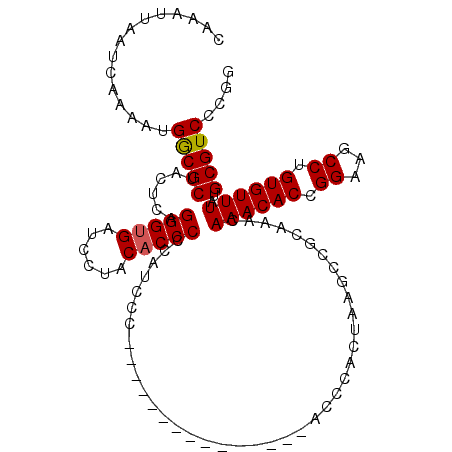
>3R_DroMel_CAF1 17083026 95 + 27905053 AACUGGCCAAUAUGGUGUGCGCAUAAUGAAUUUAUCAAAUUAAUCAAAAUGGCGCUACUCAGGGUGAUCCUACACCCCCAUCCC---------------ACCCACUAAGC ...(((.....((((.((((.(((..(((..(((......))))))..)))))))......(((((......)))))))))...---------------..)))...... ( -19.70) >DroSec_CAF1 34589 95 + 1 AACUGGCCAAUAUGGUGUGCGCAUAAUGAAUUUAUCAAAUUAAUCAAAAUGGCGCUACUCAGGGUGAUCCUACACCCCCAUCCC---------------ACCCACUAAGC ...(((.....((((.((((.(((..(((..(((......))))))..)))))))......(((((......)))))))))...---------------..)))...... ( -19.70) >DroSim_CAF1 34532 95 + 1 AACUGGCCAAUAUGGUGUGCGCAUAAUGAAUUUAUCAAAUUAAUCAAAAUGGCGCUACUCAGGGUGAUCCUACACCCCCAUCCC---------------ACCCACUAAGC ...(((.....((((.((((.(((..(((..(((......))))))..)))))))......(((((......)))))))))...---------------..)))...... ( -19.70) >DroEre_CAF1 35176 95 + 1 AACGGGCCAAUAUGGUGUGUGCAUAAUGAAUUUAUCAAAUUAAUCAAAAUGGCGCUACUAAGGGGGAUCCUACACCCCCAUCCC---------------ACCCACUAAGC ...(((......(((((.((((....(((..(((......)))))).....))))))))).(((((........))))).....---------------.)))....... ( -24.10) >DroWil_CAF1 56585 73 + 1 AGCACGCCAAUAUGGUGUGCACAUAAUGAAUUUAUCAAAUUAAUCAAAAUGGCACUGCAAAGAGUGUCCUUAC------------------------------------- .(((((((.....)))))))..............................((((((......)))))).....------------------------------------- ( -19.50) >DroYak_CAF1 40982 110 + 1 AACUGGCCAAUAUGGUGUGCGCAUAAUGAAUUUAUCAAAUUAAUCAAAAUGACGCUACUCGGGGUGAUCCCACACCCUUAUCCCCAUCCUACAACCCACCCCCACUAAAC ............(((((.((((((..(((..(((......))))))..))).))).....((((((..............................)))))))))))... ( -19.01) >consensus AACUGGCCAAUAUGGUGUGCGCAUAAUGAAUUUAUCAAAUUAAUCAAAAUGGCGCUACUCAGGGUGAUCCUACACCCCCAUCCC_______________ACCCACUAAGC .....(((.....)))((((.(((..(((..(((......))))))..)))))))......(((((......)))))................................. (-12.31 = -13.07 + 0.75)



| Location | 17,083,061 – 17,083,160 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17083061 99 + 27905053 CAAAUUAAUCAAAAUGGCGCUACUCAGGGUGAUCCUACACCCCCAUCCC---------------ACCCACUAAGCCGCAAACAAACACCGGAAGCCUGUGUUUAUGCGUCCCGG ...............(((((......(((((......))))).......---------------..................((((((.((...)).))))))..))))).... ( -20.80) >DroSec_CAF1 34624 99 + 1 CAAAUUAAUCAAAAUGGCGCUACUCAGGGUGAUCCUACACCCCCAUCCC---------------ACCCACUAAGCCGCAAACAAACACCGGAAGCCUGUGUUUAUGCGUCCCGG ...............(((((......(((((......))))).......---------------..................((((((.((...)).))))))..))))).... ( -20.80) >DroSim_CAF1 34567 99 + 1 CAAAUUAAUCAAAAUGGCGCUACUCAGGGUGAUCCUACACCCCCAUCCC---------------ACCCACUAAGCCGCAAACAAACACCGGAAGCCUGUGUUUAUGCGUCCCGG ...............(((((......(((((......))))).......---------------..................((((((.((...)).))))))..))))).... ( -20.80) >DroEre_CAF1 35211 99 + 1 CAAAUUAAUCAAAAUGGCGCUACUAAGGGGGAUCCUACACCCCCAUCCC---------------ACCCACUAAGCCGCAAACAAACACCGGAAGCCUGUGUUUAUGCGUCCCGG ...............(((((......(((((........))))).....---------------..................((((((.((...)).))))))..))))).... ( -23.80) >DroYak_CAF1 41017 114 + 1 CAAAUUAAUCAAAAUGACGCUACUCGGGGUGAUCCCACACCCUUAUCCCCAUCCUACAACCCACCCCCACUAAACAGCAAACAAACACCGGAAGCCUGUGUUUAUGCGUCCCGG ...............(((((.....((((((..............................))))))...............((((((.((...)).))))))..))))).... ( -22.91) >consensus CAAAUUAAUCAAAAUGGCGCUACUCAGGGUGAUCCUACACCCCCAUCCC_______________ACCCACUAAGCCGCAAACAAACACCGGAAGCCUGUGUUUAUGCGUCCCGG ...............(((((......(((((......)))))........................................((((((.((...)).))))))..))))).... (-20.06 = -20.10 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:36 2006